Witkop classification
Modified classification
OMIM #
Gene
TYPE I hypoplastic
IA – hypoplastic pitted (AD)
AD generalized hypoplastic
?
LAMB3
IB – hypoplastic local (AD)
AD generalized hypoplastic
104500
ENAM
IC – hypoplastic local (AR)
AR local hypoplastic
204650
ENAM
ID – hypoplastic smooth (AD)
AD generalized thin hypoplastic
?
?
IE – hypoplastic smooth (X linked)
X-linked generalized thin hypoplastic
301200
AMELX
IG – enamel agenesis (AR)
AR generalized thin hypoplastic (enamel-renal syndrome)
614253
FAM20A
204690
TYPE II hypomaturation
IIA – pigmented hypomaturation (AR)
AR pigmented hypomaturation
(IIA1) 204700
MMP20
(IIA2) 612529
KLK4
(IIA3) 613211
WDR72
(IIA4) 614832
C4ORF
IIB – hypomaturation (X linked)
X-linked hypomaturation
301200
AMELX
IIC – snowcapped (X linked)
X-linked snowcapped
301200
AMELX
TYPE III hypocalcified
IIIA – hypocalcified (AD)
AD hypocalcified
130900
FAM83H
IIIB – hypocalcified (AR)
AR hypocalcified
Not listed
SLC24A4
TYPE IV hypomaturation-hypoplastic with taurodontism
IVA – hypomaturation-hypoplastic with taurodontism (AD)
(AD) Tricho-dento-osseous syndrome
AI 104530
DLX3
TDO 190320
IVB – hypomaturation-hypoplastic with taurodontism (AR)
?
?
?
Inheritance of Amelogenesis Imperfecta
The early reports of enamel defects identified that they could be inherited by all the known modes of Mendelian inheritance: autosomal dominant, autosomal recessive and X-linked recessive [9]. The prevalence of the different subtypes varies with autosomal recessive AI types appearing to have a higher prevalence compared with the autosomal dominant AI types in Middle Eastern countries [10]. The presence of a family history of an enamel defect is very helpful in diagnosing whether the condition is indeed AI and what the AI subtype might be. Clinicians should ask patients and parents if anyone in the family has a similar enamel defect. If so, which family members and was there male-to-male transmission suggesting autosomal dominant inheritance. Were males and females similarly affected or did females often show a decreased level of severity? In the case of autosomal inherited traits, males and females will typically be similarly affected. However, if the trait is X-linked, males will typically have more severe manifestations compared with females due to lyonization [23]. Female cells only express one of their X chromosomes in any given cell so they will present a mosaic profile of affected and unaffected cells when they carry a mutation on a gene that resides on their X chromosome. Thus, enamel in females can be mildly or severely affected depending on the number of ameloblasts expressing DNA from the X chromosome with or without the mutant gene. This phenomenon of lyonization is clearly seen in the different X-linked AI subtypes where female enamel may be only mildly hypoplastic or variable in color changes that can present as streaks down the enamel of the tooth crown (Fig. 5.1) [24].
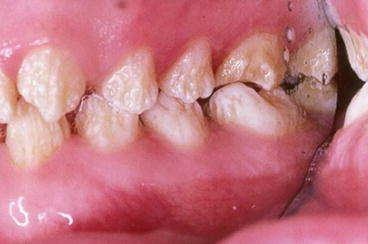

Fig. 5.1
All mutations in the AMELX gene that cause premature stop codons and loss of the C-terminus of the protein cause hypoplastic enamel. Females with these mutations will have variable areas of thick and thin enamel as seen in this affected female’s permanent dentition
The clinical phenotypes seen with the different genetic mutations are markedly different. These phenotypic variations occur for several different reasons. There are multiple genes now known to cause AI and these different genes code for proteins that have diverse functions. Thus, it is not surprising that mutations in the amelogenin gene that codes for the most abundant enamel matrix protein (amelogenin) would be associated with a very different phenotype compared with mutations in a gene coding for a proteinase like MMP20 that is involved in processing the enamel matrix proteins. There are now over 10 different genes coding for different AI types as reviewed in Table 5.1 [19, 25–27]. These genes code for proteins that are part of the extracellular matrix, proteinases that process the matrix, proteins involved in cellular attachments, proteins involved in ion regulation, genes that regulate the expression of other genes and genes whose protein functions remain unknown at this time (Table 5.2).
Table 5.2
Amelogenesis imperfecta genes and function
|
Gene
|
Protein
|
Functiona
|
|---|---|---|
|
AMELX
|
Amelogenin
|
Most abundant enamel matrix protein (90 %) important in enamel structure formation
|
|
ENAM
|
Enamelin
|
Enamel matrix protein important in crystallite elongation
|
|
WDR72
|
WD repeat domain 72
|
Intracellular protein may involve vesicle turnover
|
|
FAM83H
|
Family with sequence similarity 83, member H
|
Intracellular protein may involve keratin interaction
|
|
MMP20
|
Matrix metalproteinase 20
|
Removal of enamel matrix during secretory stage
|
|
KLK4
|
Kalikrien 4
|
Removal of enamel matrix during maturation stage
|
|
C4ORF26
|
Chromosome 4 open reading frame 26
|
Extracellular phosphoprotein with capacity to promote nucleation of mineral
|
|
DLX3
|
Distal-less 3 protein
|
Transcription factor
|
|
LAMB3
|
Laminin 5 beta chain
|
Cell attachment
|
|
SLC24A4
|
Solute carrier family 24 group4
|
Calcium transporter
|
Hypoplastic Amelogenesis Imperfecta
The primary feature of the different types of hypoplastic AI is some degree of thinning of the enamel. The currently known genes associated with this phenotype include AMELX, ENAM, LAMB3, and FAM20A. Currently all the AMELX mutations causing loss of protein due to signal peptide changes or loss of the C-terminus of the amelogenin protein result in a generalized thinning of the enamel layer in males and variable thinning in females (Fig. 5.1). Females with these types of AMELX mutations will have vertical furrows and grooves affecting the enamel thickness [20]. The difference in phenotype between males and females with X-linked AI is due to lyonization as described previously.
Many of the families identified as having autosomal recessive generalized hypoplastic amelogenesis imperfecta (OMIM #614253 AI and gingival fibromatosis syndrome) and mutations in the FAM20A gene have been shown to have microcalcifications in their kidneys and thus have enamel-renal syndrome (OMIM#204690) [28–30]. This condition has been reported using a variety of names with the typical oral phenotype including generalized thin hypoplastic enamel, failure of eruption of teeth and gingival enlargement (Fig. 5.2a, b). Management of the dentition can be very difficult due to the combination of these features and the rather common occurrence of resorption of the unerupted teeth.
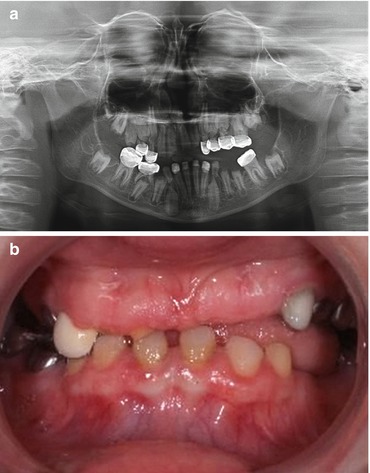

Fig. 5.2
(a) AR generalized thin hypoplastic AI with a failure of some teeth to erupt as seen in this 10-year-old females panoramic radiograph result from FAM20A mutations. (b) Clinically the teeth were small and spaced due to the very thin enamel, and the gingiva was fibrotic and overgrown in the maxillary anterior region. This affected individual also had multiple small calcifications in her kidneys consistent with the diagnosis of enamel-renal syndrome
Mutations in the ENAM gene are associated with two distinct phenotypes. Mutations that cause a loss of protein are thought to result in a localized pitted phenotype due to haploinsufficiency (only a single functional copy of a gene) where a greatly diminished amount of protein is produced by the ameloblasts because only one of the two allelic ENAM genes produces a functional protein [31, 32]. This phenotype (OMIM#104500) is inherited as an autosomal dominant trait and is characterized by horizontal pitting or grooves that decorate the clinical crown. These hypoplastic pitted/grooved areas are juxtaposed between areas of enamel that are full thickness. The enamel color appears normal indicating a normal mineral content. Other ENAM mutations produce a generalized thin hypoplastic phenotype (OMIM#104500) through a dominant negative mechanism where it is thought that some dysfunctional protein is produced that interferes with the normal process causing a more global amelogenesis dysfunction and thin enamel over the entire dental crown. There are a few case reports of hypoplastic AI due to ENAM mutations being inherited through an autosomal recessive manner (OMIM#204650) [33].
Some presentations of AI have been shown to be associated with mutations that have effects in other tissues. Hypoplastic AI has recently been associated with mutations in genes that are important in cell-to-cell attachments such as LAMB3 [27]. The protein product from the LAMB3 gene and two other genes form the protein laminin 5 that is critical for anchoring the epidermis and dermis. Some mutations in this gene are associated with junctional epidermolysis bullosa (OMIM# 226700, 226650) that is associated with tissue fragility and a propensity to develop large blisters and a hypoplastic pitted to generalized thin enamel phenotype [34]. Recently families with no predilection or history of blistering or skin manifestations have been identified to have LAMB3 gene mutations associated with a hypoplastic enamel phenotype. It is likely that other genes important in cell-cell or cell-matrix interactions or other ameloblast critical functions will be found to be associated with hypoplastic enamel defects in the future. The majority of enamel defects reported with syndromes are of the hypoplastic phenotype although many of these conditions have not been fully characterized to determine if hypomineralization also is a phenotypic component.
Hypomaturation Amelogenesis Imperfecta
Stay updated, free dental videos. Join our Telegram channel

VIDEdental - Online dental courses


