Biofilm and Periodontal Microbiology
Wim Teughels, Christof Godts, Marc Quirynen and Nick Jakubovics
The human fetus inside the uterus is sterile, but, as soon as it passes through the birth canal, it acquires vaginal and fecal microorganisms.79,107 Within 2 weeks, a nearly mature microbiota is established in the gut of the newborn baby. After weaning (>2 years), the entire human microbiota is formed and comprises a very complex collection of hundreds of different types of bacteria that totals approximately 1014 microbial cells.249 From this moment on, our body contains 10 times more bacteria than human cells. It has been estimated that, for a normal, healthy human being, the bacterial population comprises 2 kg of the total body weight. This is fascinating if one realizes that the average human brain weighs only about 1.4 kg.
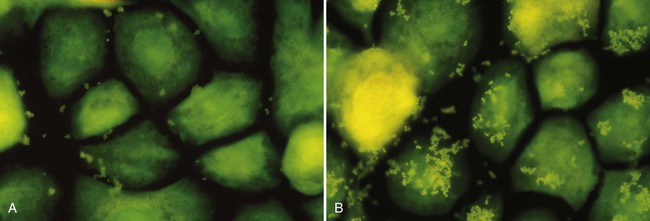
The colonization of the oral cavity also starts close to the time of birth (Table 8-1). Within hours after birth, the sterile oral cavity will be colonized by low numbers of mainly facultative and aerobic bacteria.369 At that time, the oral microbiota of newborns closely resembles the mother’s vaginal microbiota or, for newborns delivered by cesarean section, the mother’s skin microbiota.79 From the second day, anaerobic bacteria can be detected in the infant’s edentulous mouth.92,319 The number of oral bacteria increases gradually as a result of exposure to external environmental microbial sources.179,272,319 Streptococcus salivarius and Streptococcus mitis (Figure 8-1, A) have been identified as the first and most dominant oral microbes to colonize the oral cavity of newborn infants.177,178,199Veillonella spp. (Figure 8-1, B), Neisseria spp., Actinomyces spp. (Figure 8-1, C and D), and Staphylococcus spp. are also among the first colonizers of the oral cavity. After tooth eruption, a more complex oral microbiota is established. The species that colonize the teeth after eruption include Streptococcus sanguinis (Figure 8-1, D), Lactobacillus spp. (Figure 8-1, E), and Streptococcus oralis. Oral streptococci, including S. oralis, Streptococcus anginosus, mutans streptococci (Streptococcus mutans and Streptococcus sobrinus), and Streptococcus gordonii (Figure 8-1, F) are commonly reported to be present after the first year of life.49,53,219,272 In addition, anaerobes, including Fusobacterium spp. (Figure 8-1, G) and Prevotella spp. (Figure 8-1, H and I), can also be detected in young children.49,179 In later childhood, the bacterial diversity and numbers in the oral cavity increase as more teeth erupt and provide more areas for the adherence and retention of bacteria.33 Because of the paucity of longitudinal studies, relatively little is known about the initial colonization of key microbes found in the oral cavity of children and adults.199 It is estimated that more than 500 different species are capable of colonizing the adult mouth and that any individual typically harbors around 50 to 150 different species.25,252,364 When one thinks about bacteria, one almost immediately associates them with different pathologies. However, most oral bacteria are harmless commensals under normal circumstances. This means that this microbiota lives in harmony with its host but that, under specific conditions (i.e., increased mass and/or pathogenicity, suppression of commensal or beneficial bacteria, and/or reduced host response), disease can occur. The importance of the commensal microbiota is clearly illustrated by the development of yeast infections when the normal oral microbiota is reduced, such as after a longer period of systemic antibiotic usage.407 In addition, it has been shown that aggressive periodontitis is associated with a loss of colonization of S. sanguinis.372 On the other hand, it was recently shown in mice that the commensal microbiota is required for P. gingivalis–induced bone loss.2,72

The Oral Cavity From a Microbe’s Perspective
The ability of a bacterium to adhere to its host is crucial for the induction of infectious diseases, such as gingivitis or periodontitis.310 Oral bacteria and especially pathogenic bacteria, such as Porphyromonas gingivalis (Figure 8-1, H and J) and Aggregatibacter actinomycetemcomitans (Figure 8-1, K), have a large battery of virulence factors, one of which is the ability to adhere to hard intraoral surfaces and/or to the oral mucosae (Figure 8-2).56,58,105,358
• The intraoral and supragingival hard surfaces (teeth, implants, restorations, and prostheses)
• Subgingival regions adjacent to a hard surface, including the periodontal/periimplant pocket (characterized by the presence of crevicular fluid, the root cementum or implant surface, and the pocket epithelium)
• The buccal palatal epithelium and the epithelium of the floor of the mouth
In health, there is a core set of microorganisms that are almost universally present in these ecosystems. This core microbiome includes members of the phyla Firmicutes (Streptococcus spp., Veillonella spp., and Granulicatella spp.), Proteobacteria (Neisseria spp., Campylobacter spp., and Haemophilus spp.), Actinobacteria (Corynebacterium spp., Rothia spp., and Actinomyces spp.), Bacteroidetes (Prevotella spp., Capnocytophaga spp., and Porphyromonas spp.) and Fusobacteria (Fusobacterium spp.).149,200,433
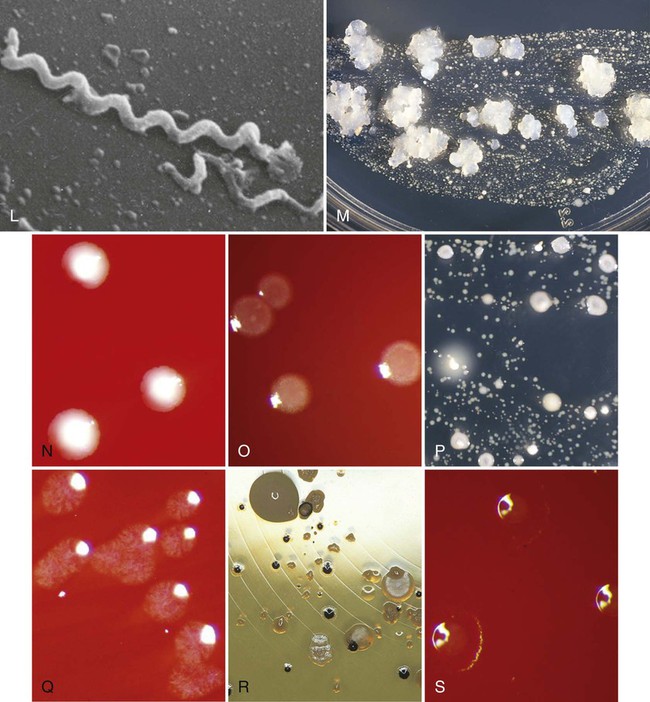
Table 8-2 summarizes several publications that discuss the detection frequency of periodontopathogens in these different niches. Most species (with the exception of spirochetes) (Figure 8-1, L) are able to colonize all of them. Some periodontopathogens (e.g. Fusobacterium nucleatum [Figure 8-1, G], Prevotella intermedia [Figure 8-1, H]) are involved in the etiology of tonsillitis, and most periodontopathogens are able to colonize the maxillary sinus.37,404
TABLE 8-2
Intraoral Habitats (Periodontal Pockets, Buccal Mucosa, Tongue, Saliva, Tonsils, and Supragingival Plaque) for Periodontopathogenic and Cariogenic Species
| DIFFERENT INTRAORAL NICHES | ||||||||||
| Authors | Infection | Age (n) | Species | No. of Positive Patients | Periodontal Pockets | Buccal Mucosa | Tongue | Saliva | Tonsils | Supragingival Plaque |
| Asikainen et al, 1991 | Periodontitis | Adult | A.a. | — | 100%* | — | 56% | 72% | — | — |
| Petit et al, 1994 | — | Child | A.a. | 5 | 4† | 3 | 1 | 3 | 2 | — |
| (45) | P.g. | 1 | 0 | 1 | 0 | 1 | 1 | — | ||
| P.i. | 34 | 23 | 18 | 25 | 31 | 22 | — | |||
| Spi | 13 | 6 | 0 | 7 | 3 | 1 | — | |||
| Petit et al, 1994 | Periodontitis | Adult | A.a. | 13 | 13† | 11 | 11 | 11 | 5 | — |
| (24) | P.g. | 18 | 18 | 14 | 7 | 10 | 11 | — | ||
| P.i. | 24 | 24 | 22 | 23 | 23 | 23 | — | |||
| Spi | 22 | 22 | 0 | 2 | 5 | 1 | — | |||
| von Troil-Lindén et al, 1995 | Periodontitis | Adult | A.a. | — | 6‡ | — | — | 6 | — | — |
| (10) | P.g. | — | 7 | — | — | 4 | — | — | ||
| P.i. | — | 10 | — | — | 9 | — | — | |||
| Danser et al, 1994 | Perio/E | Adult | A.a. | 2 | 2/—§ | 2/0 | 2/0 | 2/0 | 2/0 | 1/0 |
| (8) | P.g. | 6 | 6/— | 2/0 | 4/0 | 4/0 | 3/0 | 2/0 | ||
| P.i. | 8 | 8/— | 4/1 | 6/3 | 6/4 | 5/3 | 6/1 | |||
| Prev. | 8 | 3/— | 4/5 | 7/6 | 7/5 | 4/6 | 5/4 | |||
| Danser et al, 1996 | Perio/R | Adult | A.a. | 11 | 9/4|| | 10/9 | — | 5/2 | — | 2/0 |
| (15) | P.g. | 10 | 6/5 | 9/7 | — | 4/1 | — | 6/0 | ||
| P.i. | 15 | 11/1 | 15/15 | — | 14/13 | — | 11/1 | |||
| Gibbons and van Houte, 1975 | —¶ | —¶ | S.m. L. |
— | ? | <1|| | <1 | <1 | — | 0 to 50 |
| ? | <0.1 | < 0.1 | <1 | — | 0 to 1 | |||||
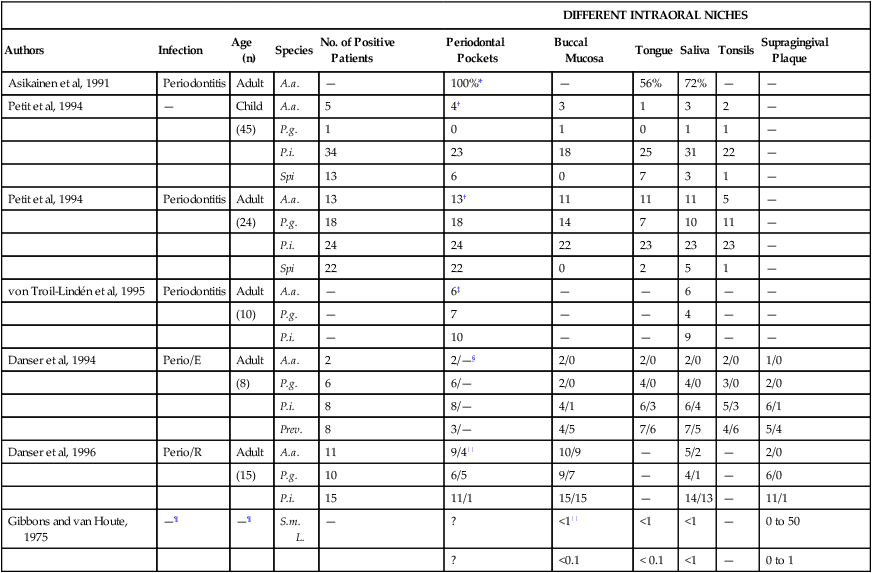
*Percentage of “specific” positive sites in positive patients.
†Number of “specific” positive sites in positive patients.
‡Number of “specific” positive sites in patients with advanced periodontitis.
§Number of “specific” positive sites in positive patients before/after full mouth extraction (E) (e.g., 2 patients were positive for A.a. before extraction, and no patients were positive for A.a. after full mouth extraction).
||Number of “specific” positive sites in positive patients before/after periodontal therapy including surgery (R) (e.g., 9 patients were positive for A.a. before periodontal therapy as compared with only 4 patients after periodontal therapy).
¶Percentage of total flora cultivable in anaerobically incubated agar in nonspecific patients as estimated from several studies.
The soft-tissue surfaces are actively involved in the process of bacterial adhesion and colonization.146 They employ a variety of mechanisms to prevent the adhesion of pathogenic organisms, with shedding being one of the most important. The high turnover rate of the intraoral epithelial cells, especially of the gingiva, prevents the permanent accumulation of large masses of microorganisms on these surfaces. In essence, this is a natural cleansing mechanism. However, bacteria can also adhere to host cells and form a commensal relationship that is beneficial for both parties.
The host vaginal epithelial cells, for example, supply glucose for the colonized lactobacilli, which in turn produce acid. A lowering of the pH prevents the growth of many other species that have deleterious effects on the vaginal environment.317 As such, these endogenous bacteria and their products can be considered necessary and beneficial components of a healthy body.
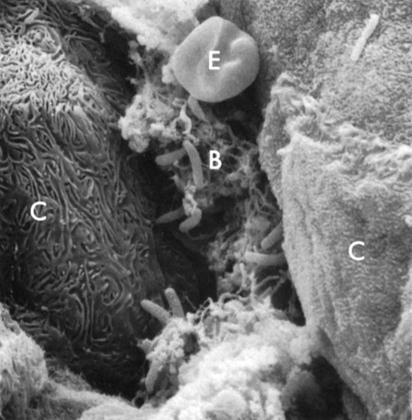
In periodontal pockets, studies have shown high numbers of bacteria attached to pocket epithelial cells in vivo. Areas of gingival inflammation are characterized by an increased number of adhering bacteria.82,392 These adhering bacteria can also infiltrate the pocket wall in relatively large numbers and reach the underlying stroma (Figure 8-3).99,232,327 In general, there is a positive correlation between the adhesion rate of pathogenic bacteria to different epithelia and the susceptibility of the affected patient to certain infections.263
Women prone to urinary tract infections, for example, harbor five times more bacteria per cell in adhesion assays of Escherichia coli to different epithelial cells of their urogenital tract (periurethral, vaginal, or uroepithelial cells). Similar observations have been made regarding the adhesion of Streptococcus pneumoniae to nasopharyngeal epithelial cells of children prone to recurrent otitis media infections as well as regarding the adhesion of Haemophilus influenzae to buccal cells of subjects prone to acute bronchitis.69,381
There are some indications that the same may be true for periodontal infections. Isogai and coworkers reported a significantly lower adherence rate of P. gingivalis and P. intermedia strains to gingival epithelial cells in rats that were resistant to gingivitis compared with susceptible rats.155 An in vitro study of cultured human pocket epithelial cells (Figure 8-4) showed a similar tendency when patients who were resistant to periodontitis were compared with patients with severe periodontal breakdown.299
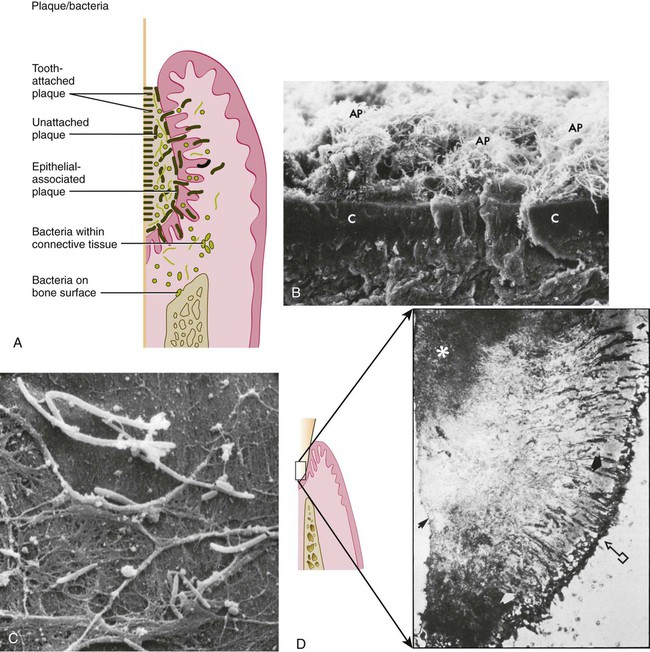
In the periodontal pocket, different strategies contribute to bacterial survival, such as adhesion to the pocket epithelium and, when dentine is encountered, the colonization of the dentine tubules.290 The crevicular fluid with its constant outflow does not favor the maintenance of unattached bacteria in the periodontal pocket.
It has been suggested that teeth are the primary habitat for periodontopathogens, because soon after a full-mouth tooth extraction in patients with severe periodontitis, key pathogens such as A. actinomycetemcomitans and P. gingivalis disappeared from the oral cavity as determined by bacterial culturing techniques.71 Prevotella intermedia and other black-pigmented Prevotella spp. did remain, but at lower detection frequencies and numbers (see Table 8-2).
The same applies to edentulous infants and wearers of full dentures in whom significant proportions of periodontopathogens have been recorded, with the exception of A. actinomycetemcomitans and P. gingivalis.70,179 Therefore, teeth were considered as a “porte d’entrée” for periodontopathogens.
However, studies involving the use of molecular tools to detect and quantify oral bacteria seem to indicate that A. actinomycetemcomitans and P. gingivalis are not entirely eradicated after full-mouth extraction. They may remain colonizers of the oral cavity, but, when teeth are lost, their relative numbers decrease.304
Alternatively, cariogenic species were thought to be relatively restricted to solid surfaces (see Table 8-2). Therefore, S. mutans (Figure 8-1, M) was often considered an obligate periphyte.380 In some studies, this species was only detected from the time that the deciduous teeth erupted in the oral cavity.50 In a longitudinal observation of adults with severe dental caries, the cariogenic species fell below detection level after full-mouth extraction but reappeared a few days after denture insertion.51 On the basis of these reports and on their own observations, Caufield and Gibbons concluded that most of the S. mutans cells in the saliva or on the tongue are derived from the biofilm present on the teeth and that the mucosae could not act as a reservoir for the infection of teeth by those organisms.54 They suggested a “window of infectivity” for the acquisition of S. mutans at a mean age of 26 months (range, 9 to 44 months).53 This observation has been supported by a few clinical studies, which showed that the initial colonization of S. mutans varied between 7 to 36 months, which is the time period that coincides with the eruption of the primary teeth.7,49,101 On the other hand, longitudinal studies by Wan and coworkers408,409 and Law and Seow198 showed that S. mutans colonization increased with the age of children, without any discrete window of infectivity. There is now clinical evidence that S. mutans can be detected in the mouths of predentate children, before the eruption of the first tooth.243,408,410
Bacteria and Their Biofilm Mode of Living
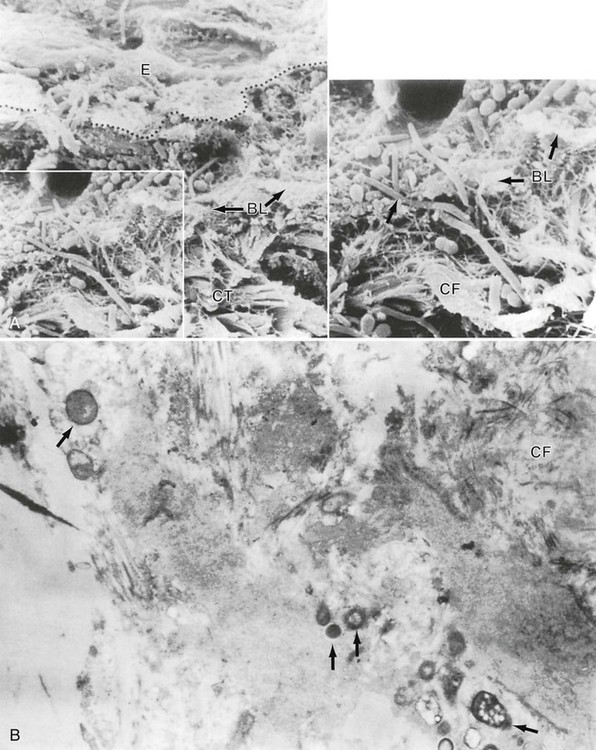
The importance of surfaces for microbial growth was recognized as early as the 1920s, when a number of workers independently noted that bacteria growing on glass slides submerged in soil were different from those that could be cultured in broth.197 However, it was not until around 50 years later that sessile microbial populations were considered to be sufficiently different from free-living microorganisms to merit their own name, and the term biofilms was coined (Figure 8-5 and Figure 8-6, B and C). Biofilms are composed of microbial cells encased within a matrix of extracellular polymeric substances, such as polysaccharides, proteins, and nucleic acids.
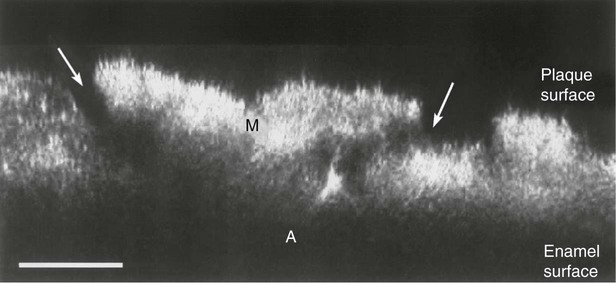
Biofilm bacteria are often up to 1000 times more resistant to antimicrobial agents than their planktonic counterparts.9,92 Bacteria that grow in multispecies biofilms interact closely with neighboring cells. Sometimes these interactions are mutually beneficial, as is the case when one organism removes another’s waste products and uses them as an energy source. In other instances, bacteria compete with their neighbors by secreting antibacterial molecules such as inhibitory peptides (bacteriocins) or hydrogen peroxide (H2O2). In addition, the biofilm mode of growth facilitates cell–cell signaling and deoxyribonucleic acid (DNA) exchange between bacteria. It is clear that microbial ecology within biofilm communities is highly complex and that, in many cases, knowledge is only emerging at this point.
Biofilms are heterogeneous: variations in biofilm structure exist within individual biofilms and between different types of biofilms. However, a number of structural features that are common to many biofilms have been noted. For example, biofilms frequently contain microcolonies of bacterial cells. Water channels are commonly found in biofilms, and these can form a primitive circulatory system that removes waste products and brings fresh nutrients to the deeper layers of the film. Surface structures, such as fronds, can dissipate the energy of fluid flowing over the biofilm and lead to the rapid blockage of vessels.81 In mixed-species biofilms, there is often heterogeneity in the distribution of different species. Steep chemical gradients exist, such as those of oxygen or pH, and these produce distinct microenvironments within the biofilm.
Microbial populations on the surfaces of teeth (dental plaque) are excellent examples of biofilm communities (Figure 8-7). The architecture of a dental plaque biofilm has many features in common with other biofilms. It is heterogeneous in structure, with clear evidence of open fluid-filled channels running through the plaque mass64,65,425 (Figure 8-5). Nutrients make contact with the sessile (attached) microcolonies by diffusion from the water channels to the microcolony rather than from the matrix. The bacteria exist and proliferate within the intercellular matrix through which the channels run. The matrix confers a specialized environment that distinguishes the bacteria that exist within the biofilm from those that are free-floating; this is the so-called “planktonic state” in solutions such as saliva or crevicular fluid. The biofilm matrix functions as a barrier. Substances produced by bacteria within the biofilm are retained and concentrated, which fosters metabolic interactions among the different bacteria.
Organic constituents of the matrix include polysaccharides, proteins, glycoproteins, lipid material, and DNA.215 Albumin, which probably originates from crevicular fluid, has been identified as a component of the plaque matrix. The lipid material consists of debris from the membranes of disrupted bacterial and host cells, bacterial vesicles, and possibly food debris. Glycoproteins from the saliva are an important component of the pellicle that initially coats a clean tooth surface, but they also become incorporated into the developing plaque biofilm. Polysaccharides produced by bacteria also contribute to the organic portion of the matrix. They play a major role in maintaining the integrity of the biofilm.
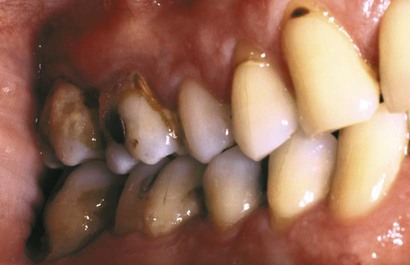
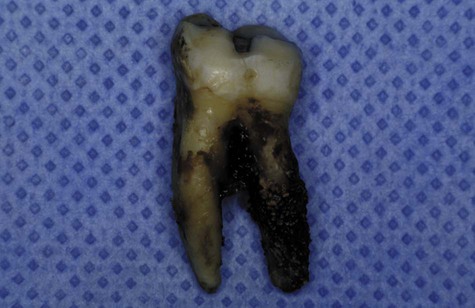
The inorganic components of plaque are predominantly calcium and phosphorus, with trace amounts of other minerals such as sodium, potassium, and fluoride. The source of inorganic constituents of supragingival plaque is primarily saliva. As the mineral content increases, the plaque mass becomes calcified to form calculus (Figure 8-8). Calculus is frequently found in areas of the dentition adjacent to salivary ducts (e.g., the lingual surface of the mandibular incisors and canines, the buccal surface of the maxillary first molars), which reflects the high concentration of minerals available from saliva in those regions. The inorganic components of subgingival plaque are derived from crevicular fluid (a serum transudate). The calcification of subgingival plaque also results in calculus formation (Figure 8-9). Subgingival calculus is typically dark green or dark brown, which probably reflects the presence of blood products that are associated with subgingival hemorrhage.
Structure of a Mature Dental Plaque Biofilm
Dental plaque (see Figure 8-7) is defined clinically as a structured, resilient, yellow-grayish substance that adheres tenaciously to the intraoral hard surfaces, including removable and fixed restorations.30 The tough extracellular matrix makes it impossible to remove plaque by rinsing or with the use of sprays. Plaque can thus be differentiated from other deposits that may be found on the tooth surface, such as materia alba and calculus. Materia alba refers to soft accumulations of bacteria, food matter, and tissue cells that lack the organized structure of dental plaque and that are easily displaced with a water spray. Calculus is a hard deposit that forms via the mineralization of dental plaque and that is generally covered by a layer of unmineralized plaque (Table 8-3).
Dental plaque is composed primarily of microorganisms. One gram of plaque (wet weight) contains approximately 1011 bacteria.340,361 The number of bacteria in supragingival plaque on a single tooth surface can exceed 109 cells. In a periodontal pocket, counts can range from 103 bacteria in a healthy crevice to more than 108 bacteria in a deep pocket. With the use of highly sensitive molecular techniques for microbial identification, it has been estimated that more than 500 distinct microbial phylotypes can be present as natural inhabitants of dental plaque.1
Any individual may harbor 150 or more different species. Nonbacterial microorganisms that are found in plaque include archaea, yeasts, protozoa, and viruses.62,206
• Supragingival plaque is found at or above the gingival margin; when in direct contact with the gingival margin, it is referred to as marginal plaque.
• Subgingival plaque is found below the gingival margin, between the tooth and the gingival pocket epithelium.
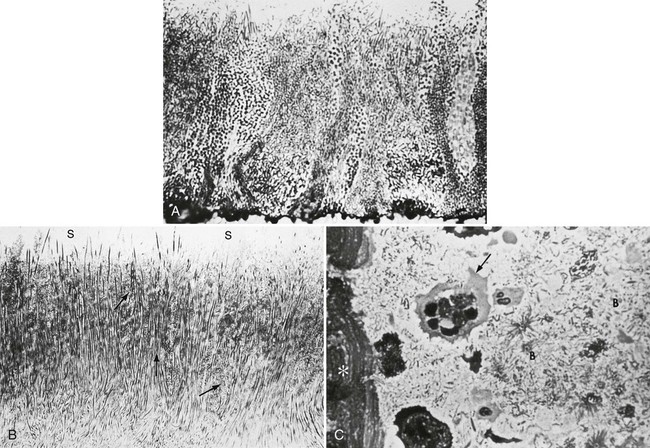
Supragingival plaque typically demonstrates the stratified organization of a multilayered accumulation of bacterial morphotypes (Figure 8-10).437 Gram-positive cocci and short rods predominate at the tooth surface, whereas gram-negative rods and filaments as well as spirochetes predominate in the outer surface of the mature plaque mass.
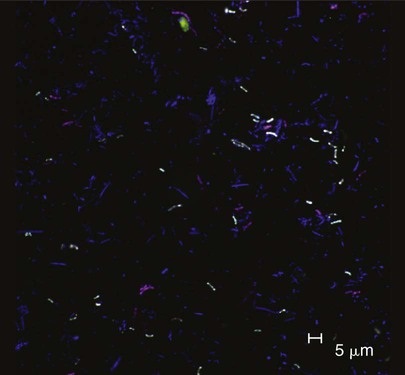
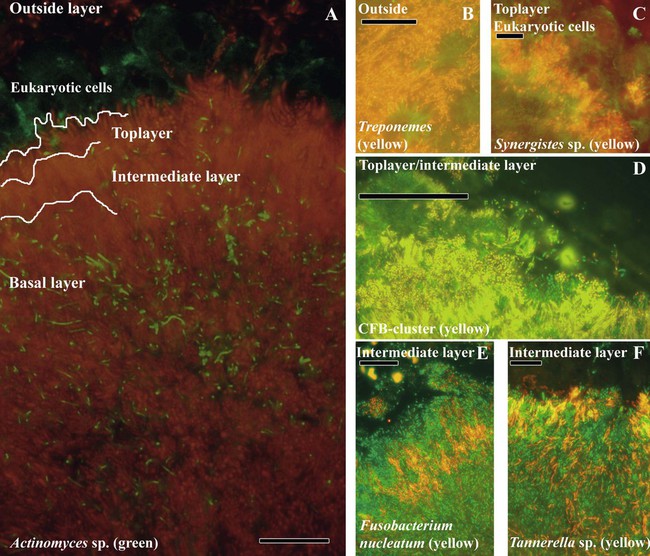
The identification of bacteria within intact dental plaque represents a significant challenge. Most of the previously cited studies of dental plaque microbiota used techniques that involved the disruption of the dental plaque matrix followed by microbial culture (Figure 8-11) or culture-independent identification. Techniques have been developed that allow for the specific visualization of individual bacteria within mixed populations. With these methods, specific labeling is achieved by using nucleic acid probes (fluorescence in situ hybridization [FISH]) or specific antibodies (immunofluorescence). FISH has been applied to identify particular species within plaque samples scraped from periodontal pockets.111,265,401 With this methodology, it is even possible to image bacteria that have never been cultured in the laboratory, such as members of the phylum Synergistetes (Figure 8-12). Intact subgingival biofilms (Figure 8-13) have also been analyzed by FISH and shown to be composed of a heterogeneous structure with organisms such as Actinomyces spp. at the base, F. nucleatum and Tannerella forsythia in the middle layers, and periodontopathogens including members of the Cytophaga–Flavobacterium–Bacteroides cluster in the outer layers.437 Supragingival biofilms have proved somewhat easier to visualize than subgingival plaque (Figure 8-14), and they have a somewhat different architecture.437 Biofilms can be cultured on retrievable enamel chips held in intraoral devices in the mouths of volunteers. The enamel pieces can then be removed and processed for FISH or immunofluorescence. Images of biofilms can be captured by confocal laser scanning microscopy, which produces three-dimensional representations of the biofilm architecture. Labeling with nucleic acid probes requires the desiccation of the sample before hybridization, and therefore some structural information may be lost. Nevertheless, this technique has been applied successfully to determine the spatial relationships between Actinomyces naeslundii and Streptococcus spp. in dental plaque.78 In these studies, A. naeslundii cells were frequently observed juxtaposed to Streptococcus spp. cells. Many strains of Actinomyces coaggregate with oral streptococci, and therefore the spatial organization of these organisms within dental plaque may be influenced by adhesin–receptor interactions.175 In fact, a protein adhesin of A. naeslundii has been shown to colocalize with a cognate oral streptococcal receptor polysaccharide in in situ biofilms using specific antibody (immunofluorescence) labeling (Figure 8-15).267 These studies clearly demonstrate that the interactions identified among bacteria isolated in the laboratory are relevant to the dental plaque biofilm.
Stay updated, free dental videos. Join our Telegram channel

VIDEdental - Online dental courses




