Genetic Susceptibility to Periodontal Disease
Scott R. Diehl, Chih-Hung Chou, Fengshen Kuo, Ching-Yu Huang and Olga A. Korczeniewska
Opportunities for advanced study designs and next-generation genomic technologies to improve the understanding of the inherited basis of periodontitis make it likely (although by no means certain) that genetic variation will become an important variable to be routinely considered by practicing dentists within the next 5 to 10 years. If and when this occurs, dentists will need to learn how to access and interpret the massive amounts of information coded in the more than 3 billion bases of DNA in the human genome and to incorporate this into improved evidence-based practices for periodontal disease prevention, diagnosis, and treatment. Alternatively, new interdisciplinary teams will need to be established with dentists as key members working closely with experts in bioinformatics, genomics, and genetic counseling so that patients will be able to reap the potentially significant benefits of these scientific advances. However the process evolves, it will surely lead to revolutionary changes in both dental education and dental practice in the not-so-distant future.8,26
Genetic and Genomic Methods of the 21st Century
Most health care professionals are aware of the many major advances in genetics accomplished during the past 20 years since the Human Genome Project officially began in 1990.46 Headlines and announcements of “breakthroughs” have appeared regularly in print and television media. Too often, many of these stories seem to promise rapid and significant improvements in health care that are totally unrealistic.11 More cautious assessments are usually not deemed as newsworthy by the media, and cautious understatement is not always in the interest of private companies and granting agencies involved in basic research and clinical trials. Consequently, the public has often received an overly optimistic picture of what the future of medical care based on genomic medicine really holds for them. Despite this all-too-common overstatement about rapid translation to clinical practice, advances in technical capabilities for genomic data acquisition and the accumulation of knowledge in the field of genetics have been truly enormous. The eventual impact of this explosion of biologic knowledge on all areas of human health, including those concerning the field of dentistry, is certain to be very substantial over the longer term.
Unfortunately, for reasons explored further in the next section, genomic advances have thus far contributed very little to advancing our understanding of the molecular–pathologic causes of periodontitis or pointed toward ways to improve treatment through “individualized” approaches that are based on a patient’s inherited genetic variation. Only recently has the possibility of making major progress been realized on the basis of newly available genomic tools and approaches involving genome-wide association studies (GWAS; pronounced “gee-wahs”) and “next-generation” DNA sequencing techniques. For these research strategies to be successful, however, they must be combined with further improvement in research definitions of periodontal disease and much larger sample sizes than have been used in most previous genetic studies of this condition. To help improve understanding, some of the commonly used terms in genetics are explained in Table 6-1.
TABLE 6-1
Glossary of Terms Relevant to the Genetics of Periodontal Disease
| Allele | One of several possible alternative forms of a gene caused by small or large differences in the DNA sequence within or near the gene. These differences arise by mutation, and some may affect the function of the gene product (i.e., a protein) or its abundance in different kinds of cells. |
| Autosome | A chromosome that is not a sex chromosome. |
| Autosomal dominant | DNA variation in a gene located on an autosome that has a dominant effect over other forms of variation at this location within the gene. When the dominant DNA sequence is present in combination with some other sequence, the gene’s function is entirely or nearly entirely determined by the dominant sequence, whereas the alternative sequence that occurs on the person’s other chromosome is essentially silent. |
| Autosomal recessive | DNA variation in a gene located on an autosome that has an effect on the gene’s function only when the person has inherited two copies: one from the mother and the other from the father. For example, if an individual has two copies of an abnormal gene that is autosomal recessive, they will be subject to the effects of that gene. |
| Chromosome | A nuclear structure that contains genetic information. Humans have 46 chromosomes that are arranged in 23 pairs. There are 22 pairs of autosomes and one pair of sex chromosomes (either XX or XY). |
| Concordance | The probability that a pair of individuals (e.g., twins) both have a certain characteristic (e.g., periodontal disease), given that one of the pair has the characteristic. Presented as a number from 0 to 1 or as a percentage. |
| Dizygotic twins | Twins that have resulted from the fertilization of two separate eggs. They are no more similar to each other (from a genetic perspective) than are nontwin siblings. Nonidentical twins. |
| Epigenetics | Term used to describe the changes in phenotype or gene expression that result from mechanisms other than changes in the underlying DNA sequences (i.e., changes in which the gene is expressed rather than a change in the DNA sequence itself). Nongenetic factors cause the organism’s genes to be expressed differently. |
| Exon | Protein coding regions of DNA. |
| Frameshift mutation | A mutation that results from the insertion or deletion of one or more nucleotides into a gene, thereby causing the coding regions to be read in the wrong frame and usually causing the protein produced to be defective in function. |
| Gene | The basic unit of heredity that occupies a specific position (locus) on a chromosome and that has specific effect(s) on the phenotype of the organism. A piece of DNA that is transcribed into a molecule of RNA and then translated into a protein. |
| Gene expression | The process by which the information in a gene is used via transcription and translation, thereby leading to the production of protein. Differences in gene expression can affect the phenotype of the organism, including the risk of disease. |
| Genetic code | In RNA and DNA, the consecutive nucleotide triplets (codons) that specify the sequence of amino acids for protein synthesis (translation). |
| Genome | The entire hereditary information of an organism. This term refers to all of the genes and other nongene portions of DNA carried by an individual cell. |
| Genotype | The genetic makeup of an organism or cell as distinct from its expressed features or phenotype. |
| Haplotype | A contraction of the term haploid genotype. This word refers to a combination of alleles at multiple loci, which are usually transmitted together on the same region of a chromosome. |
| Heredity | The passing of traits to offspring from parents or ancestors. In biology, the study of heredity is referred to as genetics. As a result of heredity, variation among individuals allows species to evolve by natural selection in response to changes in their environment or by random change over long periods of time. |
| Homozygous | The presence of identical alleles at a specific position in a gene. |
| Heterozygous | The presence of two different alleles at a specific position in a gene. |
| Intron | A DNA region within a gene that is not translated into protein. These intervening (noncoding) portions of DNA or RNA are removed during RNA processing. |
| Isoform | Any of several different forms of the same protein. Isoforms may be produced from related genes, or they may arise from the same gene via alternative splicing. Many isoforms are caused by single nucleotide polymorphisms. |
| Ligand | A molecule that binds to another molecule (usually a cellular receptor molecule). |
| Linkage | A term used to describe the tendency for certain genes to be transmitted from parent to child together because they are located close to each other on the same chromosome. |
| Linkage disequilibrium | The occurrence of specific alleles at different locations in the DNA that are relatively close to each other (linked) more often than would be expected by chance alone (disequilibrium). |
| Locus | The physical location that a gene occupies within a chromosome. (Plural: loci.) |
| Monozygotic twins | Twins with identical genetic makeup (i.e., identical twins) as a result of the fertilization of a single egg that then splits into two embryos. |
| Mutation | Changes in the DNA sequence of the genome can result from errors that occur during DNA replication or meiosis and can be caused by radiation, viruses, and mutagenic chemicals. Most mutations have very little or no measurable effect on the gene’s function; some are harmful, and a rare few may be advantageous. |
| Nucleotide | Molecules that, when linked together, make up the structural units of RNA and DNA. They are composed of a phosphate group; the bases adenine, cytosine, guanine, and thymine; and a pentose sugar. In RNA, the thymine base is replaced by uracil. |
| Penetrance | The proportion of individuals who have a particular allele/genotype who express an associated trait (phenotype). Genotypes with a high penetrance result in a larger number of individuals in the population with the associated phenotype as compared with genotypes with a low penetrance. |
| Phenotype | The observable characteristics displayed by an organism (e.g., morphology, development, gender, eye color, physiologic properties, behavior). Phenotype results from the expression of the organism’s genes as well as from the influence of environmental factors and interactions between the two. |
| Polymorphism | Polymorphism exists when two or more different phenotypes exist within different individuals of the same population. In the context of genetics, it refers to a region of the genome that varies between individual members of the population in such proportions that the rarest of them cannot be maintained just by recurrent mutation. Polymorphism may be actively maintained in populations by natural selection and also by random drift. |
| Sequencing | Determining in the laboratory the linear arrangement of nucleotides (in RNA or DNA) or amino acids (in proteins). |
| Single nucleotide polymorphism (SNP) | A polymorphism in a gene caused by a change in a single nucleotide in the DNA sequence. A large number of protein isoforms result from SNPs. SNPs occur frequently; approximately every 100 to 1000 base pairs occur as a result of deletions, insertions, and substitutions. There are estimated to be more than 10 million SNPs in the human genome. Many SNPs that occur in genes have no effect on the encoded protein, but some SNPs do influence the function of the protein that the gene produces. An SNP initially arises as a very rare mutation, but it is considered to be an SNP if it occurs in at least 1% of the population. |
| Signal transduction | A cascade of intracellular events that occurs after the binding of an extracellular signal (e.g., a hormone, a cytokine) to a receptor on the cell surface. The intracellular cascade can result in changes in gene expression in the nucleus and hence an altered phenotype of the cell (e.g., as a result of different protein production). |
| Splicing | The removal of introns from transcribed RNA. The process of removal can vary, and some exons are skipped or excluded from splicing. This causes the production of “splice variants” or “alternatively spliced” protein isoforms, thereby resulting in the formation of different proteins from the same initial RNA. |
| Transcription | RNA synthesis. The process of creating an RNA copy of an equivalent section of DNA is the first step of gene expression, and it occurs in the nucleus. The RNA copy that is produced is called messenger RNA (mRNA). |
| Translation | The first stage of protein synthesis. mRNA produced during transcription is decoded to produce an amino acid chain that will later fold into an active protein. Translation occurs in the cytoplasm: ribosomes bind to the mRNA and then facilitate decoding via the binding of transfer RNAs (tRNAs) that have complementary anticodon sequences to those of mRNA. The tRNAs carry specific amino acids that are joined together to form a polypeptide as the mRNA passes through the ribosome. |
Patterns in Populations and Pedigrees
Regrettably, the comparison of periodontitis in different populations across the globe is extremely challenging because of the lack of calibrated examiners and standardized disease definitions.6 One of the most dramatic population differences in which data quality is not an issue is the observation that both localized and generalized forms of early-onset aggressive periodontitis occur about 10 times more frequently among African Americans as compared with Caucasians.33 Human racial and ethnic groups often differ dramatically with regard to the frequency of mutations at genes that have major effects on disease risk. For example, cystic fibrosis is caused exclusively by recessive mutations in the CFTR gene, and it varies in frequency from 1 in 3000 Caucasians to 1 in 15,000 African Americans in the United States, whereas only 1 in 350,000 Japanese individuals are affected.49 It is possible that the tenfold higher prevalence of early-onset aggressive periodontitis in African Americans is caused by the elevated frequency of high-risk gene variants in this population. However, additional evidence is needed before such a conclusion can be drawn. Although comparative studies of different populations may provide clues as to possible genetic mechanisms underlying a disease, the environments of the populations may also be dissimilar in important ways. It is possible that variations in diet or in exposure to pathogenic oral bacteria or to some unknown and unmeasured environmental factors could entirely explain the observed differences in the frequency of aggressive periodontitis between population groups. Until solid data confirm a genetic basis for population differences, we need to wait before drawing firm conclusions.
Most human diseases and nondisease traits fall in the middle of this range, with heritability ranging between 0.25 and 0.75. For example, in one study, type II diabetes was estimated to have a heritability of 0.26, and abnormal glucose tolerance had a heritability of 0.61.50 Note that, for it to be feasible to use the twin method with adequate statistical power, the disease has to be fairly common so that the researcher can recruit enough twin pairs in which at least one of the twins is affected by the disease. Not surprisingly, with regard to periodontal disease, only chronic periodontitis occurs frequently enough to have been studied using the twin design. Two twin studies of modest size (i.e., 110 and 117 pairs) have been reported, and these estimate the heritability of measures of chronic periodontitis to range between 40% and 80%, thereby clearly implicating genetic variation in disease risk.43,44 Interestingly, a study of bacteria associated with periodontitis found no difference between identical versus nonidentical twins.45 This suggests (at least for these twins, most of whom did not have severe periodontitis) that inherited variation in risk is not mediated by genes that influence the presence of specific bacteria in subgingival plaque.
Another method used by genetic epidemiologists to understand and distinguish different mechanisms of transmission of diseases through families is called segregation analysis. This is relatively straightforward for traits in which mutation in a single gene causes the disease to develop with nearly 100% certainty in carriers, whereas persons who do not inherit the mutation are at little or no risk. For example, carriers of a single copy of the Huntington disease gene mutation or carriers of two copies of a cystic fibrosis gene mutation always develop these diseases if they reach the ages at which symptoms of these conditions normally emerge. By tracking the transmission of these diseases in families, it is obvious, for example, that Huntington disease is a dominant single-gene disorder: it is transmitted with 50% probability to offspring of affected individuals and thus it is often found occurring across many generations of large pedigrees. By contrast, parents of children with cystic fibrosis are very rarely affected themselves, and 25% of siblings are affected by cystic fibrosis when the disease is present in a nuclear family. This pattern of transmission is expected if a disease is recessive (i.e., it requires the inheritance of a mutated gene copy from both parents, who themselves have one normal and one mutated copy and so are not affected). For most common “complex” diseases, however, having a high-risk gene does not automatically lead to development of the disease; this phenomenon is called reduced penetrance. Furthermore, several genes or even dozens of different genes may influence disease susceptibility; this is known as oligogenic inheritance and genetic heterogeneity. Environmental exposures are also important modifiers of disease risk. Such highly complex combinations of multiple genetic and environmental risk factors make the challenge of deciphering genetic mechanisms by merely observing transmission patterns in families using the segregation analysis approach unfeasible. The limitations of this approach were humorously illustrated a number of years ago in an analysis that facetiously presented evidence of a recessive gene controlling the trait of attending medical school.39 “Risk” for this outcome among first-degree relatives of a doctor was elevated 61 times above that of the general population. More recently, a robust quantitative analysis of the family histories of characters in the Harry Potter series suggested that a dominant gene controls the inheritance of magic abilities.54 Because the etiology of periodontitis is likely to be highly complex, segregation analyses of this disease that have been reported in the literature should be viewed with considerable skepticism. Unfortunately, the simplifying assumptions required for this method make the results unreliable and potentially misleading. For highly complex diseases, such as most cases of periodontitis, assays at the DNA level need to be combined with careful evaluations of clinical measures among related individuals to derive robust conclusions about a disease’s genetic architecture. Some of the key features of the different techniques for studying the genetics of periodontal disease are explained in Table 6-2.
TABLE 6-2
Techniques for Studying the Genetics of Periodontal Disease
| Candidate gene approach | A gene-mapping approach that tests whether one allele of a gene occurs more often in patients with the disease than in subjects without the disease. These methods are also referred to as association analyses, and they aim to identify which genes are associated with the disease. Candidate genes are chosen on the basis of their known or presumed function (i.e., they have some plausible role in the disease process, such as producing a protein that is important in the disease pathogenesis). Conceptually this makes sense, but it requires some knowledge of the candidate gene to look for it! |
| Case–control studies | Studies in which the genetic makeup is compared between cases (who have the disease in question) and controls (who do not). The populations need to be carefully matched, otherwise apparent observed differences between cases and controls could arise because of ethnic or geographic variation, for example. |
| Twin studies | Comparisons of traits—including diseases in monozygotic, dizygotic, or usually both types of twins—aimed at determining whether variation in the trait among members of a population is caused by genetic variation in inherited DNA sequences, environmental exposures in the subjects’ lives, or some combination of both of these processes. Twin studies often measure the concordance rates of twins with regard to a particular trait or disease of interest. Monozygotic (identical) twins are very nearly identical in their DNA, whereas dizygotic (nonidentical) twins share an average of half of their DNA as identical sequences inherited from their parents. If a disease has high heritability, identical twins will be more likely to either be both affected or both unaffected (concordant). However, this assumption is complicated in many diseases. A genetic mutation may not have complete penetrance, and environmental conditions may contribute to the development of the disease (e.g., one twin may smoke and the other may not). Furthermore, many diseases are polygenic (i.e., caused by alterations in multiple genes). |
| Familial aggregation and relative risk | Many diseases run in families, and the degree of clustering within the family can be estimated by comparing the number of disease cases in relatives of patients to the risk of disease in the general population. Difficulties with this approach relate to the fact that, in addition to having many genes in common, family members also share many aspects of a common environment (e.g., diet, nutrition, smoking, infectious organisms, shared socioeconomic factors). |
| Segregation analyses | Statistical analyses of the patterns of transmission of a disease in families in an attempt to determine the relative likelihood that the disease is caused by a single gene with dominant or recessive inheritance, by multiple genes, or entirely by variation in exposure to risk factors. The observed proportions of offspring who have the trait or disease being evaluated (i.e., the phenotype) are compared with the proportions expected to be found in the general population. |
| Linkage analysis | A technique used to map a gene responsible for a trait to a specific location on a chromosome. These studies are based on the fact that genes that are located close to each other on the chromosome tend to be inherited together as a unit. As such, these genes are said to be “linked.” Because linkage analysis initially requires the use very expensive DNA markers, this was originally only considered justified after finding strong evidence of a genetic basis for a trait with the use of segregation analyses or family aggregation studies. One difficulty with linkage analyses is that many diseases are not caused by a single gene of “major” effect but rather by multiple genes of “minor” effect. In the latter situation, multiple genes each contribute a small amount to the phenotype, disease, or trait. The linkage study approach has little power for detection, whereas association analysis methods may still be quite powerful. |
| Genome-wide analyses | A genome-wide association study (GWAS) investigates genetic variation across the entire genome simultaneously, with the aim of identifying genetic associations related to a trait or disease of interest. The completion of the Human Genome Project in 2003 and the development of microarray technologies capable of assaying more than half a million single nucleotide polymorphisms have made GWASs possible. This method has the potential to identify the genetic contributions to common diseases. Because the entire genome is analyzed, an important advantage of this approach is that the technique permits the genetics of a disease to be investigated in a nonhypothesis-driven way. In other words, it is not necessary to correctly guess which candidate genes are most interesting to evaluate. A GWAS requires that well-characterized cases and controls be identified. A disadvantage of GWASs is that large clinical sample sizes are required to reduce the likelihood of differences between the cases and controls being observed simply by chance as a result of the hundreds of thousands of multiple statistical tests required to search the entire human genome. |
Searching for Answers in the DNA
In theory, a genetic marker can be any type of biomolecule or assay that allows us to “read” inherited differences among individuals in their DNA sequences. Blood groups, protein isozymes, and human leukocyte antigens (HLAs) were among the first developed markers, but even simple traits that are controlled by single genes (e.g., eye color) can also serve this purpose. Genomic methods have made these methods obsolete, because researchers can now determine a person’s inherited variation directly at the DNA level for a much lower cost and with greater speed and accuracy. So-called “next-generation” DNA sequencing methods are projected to enable researchers and clinicians to obtain nearly the entire 3 billion DNA base human genetic blueprint for less than $1000 a few years from now.18,67 At present, most genetic studies use a combination of whole genome arrays that can evaluate up to 1 million variable DNA sites in a single assay in combination with lower-throughput methods that are used for the fine mapping of chromosome regions of special interest.53 These regions are said to contain candidate genes of high priority for further investigation either because of the genes’ known biologic functions or because results of previous genome-wide surveys indicate strong statistical chances that disease susceptibility genes are located in certain regions of one or more chromosomes. Types of variation include single nucleotide polymorphisms (SNPs; pronounced “snips”) in which one DNA base is substituted for another; small insertions and deletions (“in/dels”) of one or more DNA bases; and larger structural changes in the DNA, such as inversions (in which a piece of DNA of hundreds or thousands of bases in size is cut out and sewn back into the chromosome in the opposite orientation) and copy number changes (in which a given segment of DNA is either missing or occurs in more than the usual two copies inherited as one copy from each parent).
Equipped with these powerful tools for rapidly measuring DNA variation, the next question to address is what kinds of study designs are most powerful for identifying the dozen or more variants that influence the risk of disease from among the millions of DNA differences that exist between any two individuals in a typical population. One method that has been highly successful for finding molecular defects related to simple genetic diseases caused by the mutation of a single gene, such as cystic fibrosis and Huntington disease, is linkage analysis.1,7 This gene-mapping strategy requires families with one or more members affected by the disease to be recruited and clinically and molecularly evaluated for a relatively small number of genetic markers. Depending on the type of marker used, as few as 500 markers or up to 10,000 markers distributed evenly across the genome are needed. Investigators usually try to recruit families with two or more close relatives, such as sibling pairs, who are affected by the disease as well as parents and other siblings who may be unaffected. With a null hypothesis that suggests that a region of the chromosome does not contain genetic variation that influences disease risk, siblings are expected to share identical genetic material inherited from their parents an average of 50% of the time. However, if the region of the chromosome being evaluated contains a gene that has a substantial effect on disease risk (e.g., increases the risk by tenfold or more), then pairs of siblings who are both affected by the disease will share the chromosome region that contains the disease gene substantially more often than 50% of the time, and the null hypothesis of 50% sharing will be statistically rejected if the study has an adequately large enough sample of such families. This simple example illustrates how linkage analysis is performed. In practice, both small and large extended families are studied; this will include the simultaneous evaluation of the sharing of genetic material among both affected and unaffected relatives. Sophisticated mathematical algorithms and computer programs are used to carry out the huge number of calculations required for data analyses.
After achieving many successes with the use of linkage analysis for “simple” diseases caused by the mutation of a single gene, this method was extended to complex diseases caused by combinations of multiple susceptibility genes and environmental risk factors. Unfortunately, these conditions proved to be beyond the reach of linkage analysis in most instances. Numerous studies conducted during the 1990s either failed to find any genes, or initially positive findings failed to replicate. Linkage studies of very large numbers of carefully diagnosed families for complex diseases (e.g., orofacial clefting, in which twin studies had firmly established heritability of 70%) identified at most a tiny fraction of this genetic variation. Mathematical analyses have subsequently shown that the linkage analysis gene-mapping strategy has extremely low statistical power for complex diseases in which each individual susceptibility gene has a relatively small effect on risk (e.g., twofold or less) and in which there is extensive heterogeneity among different families that have different combinations of susceptibility genes and environmental exposures.56 Consequently, it is not surprising that linkage analysis has only been successfully applied to syndromic forms of periodontitis; this information is summarized later in this book.
Disappointment over this setback in human disease mapping caused by the initially unrecognized limitations of linkage analysis was short-lived. An alternative approach called association analysis was also available, although this had been relegated to studies of HLA and a few other markers of special interest during the prime of linkage approaches.1,7 Mathematical analyses indicated that, if we make some reasonable assumptions about the nature of genetic factors in human disease, this method could provide adequate statistical power for finding genes of small to modest effect on risk while requiring only moderately large sample sizes that would be feasible to recruit.56 There was, however, one major catch: to search the entire genome using the GWAS method, the number of genetic markers needed was several orders of magnitude greater (i.e., 500,000 to 1 million assays needed per subject). Fortunately, advances in molecular assay technologies converged at this time period, and several methods of array-based genotyping provided such a capability at an acceptable cost.53
How association studies are used to find disease susceptibility genes is illustrated for the case–control design in Figure 6-1. Association analyses are sometimes referred to as case–control studies, although this is only one of several sampling methods that can be used (including studies of families). Genotype frequencies of an inherited DNA variant for a group of periodontitis cases are statistically compared with the frequencies of the variant in a matched group of periodontally healthy control subjects. If the genotype frequencies differ so greatly that the results are very unlikely to occur by chance, then we conclude that the genotype that is more common in the cases as compared with the controls is “associated with” increased disease risk. In Figure 6-1, 59% of the cases have the CC genotype (having inherited a C allele from both of their parents), whereas only 17% of the healthy controls inherited the CC genotype. Therefore, this DNA variant could be used to predict periodontitis risk (but not until after the finding was validated in additional independent studies). Conversely, we can also say that the AA genotype is “protective” against disease because it occurs much more often in the healthy controls (50%) as compared with periodontitis cases (8%). Ideally, the cases and healthy controls are matched as closely as possible for race/ethnicity, smoking behavior, age, gender, and so on so that differences in genotype frequency are likely to be caused by real biologic effects on disease development or progression rather than as artifacts of some kind. For example, it is well known that races and ethnic groups sometimes differ dramatically with regard to genotype frequencies as a result of their historic isolation in different geographic regions. Consider, for example, a study that had mostly Swedish cases and mostly Italian controls. We know that there are thousands of DNA variants that differ substantially among these populations because of their geographic isolation throughout human history. Few if any of these variants have anything to do with differences in disease risk, but they could falsely appear to be associated because of the failure to carefully match ethnicity in cases and controls. In practice, this is usually not a problem, provided that investigators take reasonable precautions with regard to how cases and controls are selected. It is also now routine practice to use several statistical methods to check for mismatching and then adjust for this during data analysis if it occurs.
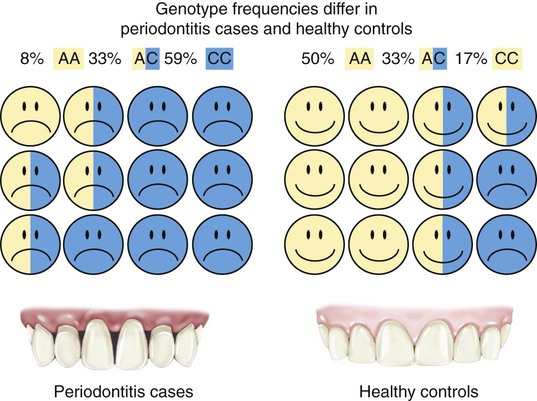
The good news is now clearly in: association studies have been a boon for the discovery of inherited genetic variation important for a wide range of complex diseases, including diabetes, cardiovascular disease, metabolic disorders, obesity, and mental illnesses. A recent review cites 24 genes identified with unquestionable statistical confidence for type 2 diabetes alone, and the list continues to grow.64 Most of these genetic polymorphisms with elevated risk are very common in the population (i.e., from 5% up to >50%). Although each variant only increases risk slightly (i.e., twofold or less), because the risk alleles are so common, they can account for a nontrivial proportion of the occurrence of disease in the population; this is a measure that epidemiologists call attributable risk.
Although great progress has been made toward understanding the etiology of many complex human diseases by using GWAS methods, the approach has nevertheless usually failed to account for most of the heritability known to exist for these conditions.12,38 A recent study found that well-established nongenetic diabetes risk factors (e.g., gender, smoking, family history, body mass index, blood lipid and glucose levels) were better predictors of risk than a combination of the top 20 genetic markers for this disease.65 To improve gene-based risk estimates, the missing heritability needs to be found. High hopes are now being placed on the emergence of next-generation DNA-sequencing tools. In theory, these may enable researchers to identify the less common (i.e., 1% to 5%) genetic variants that are predicted to have individual gene effects of greater magnitude on disease risk (i.e., greater than twofold but less than tenfold) that cannot readily be found with the use of either GWAS or linkage analysis methods.
Inherited Variation and Risk of Periodontitis
Periodontitis in Genetic Syndromes and Other Diseases
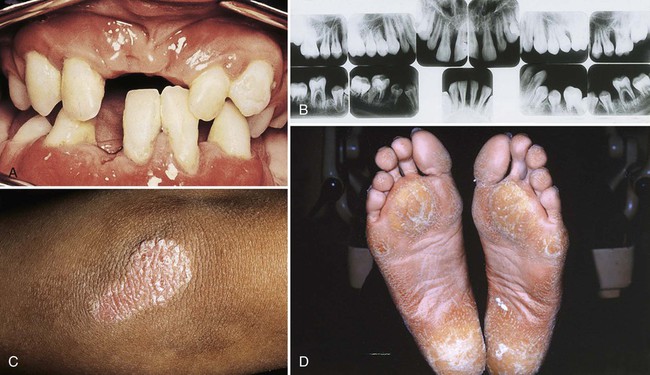
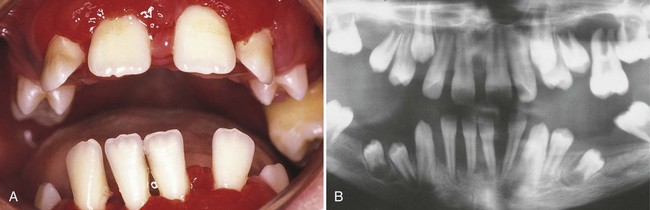
A number of extremely rare conditions consistently include periodontitis among the array of clinical manifestations that define a syndrome. Many genetic syndromes involve mutations of single genes or larger chromosomal regions. However, a number of syndromes, such as fetal alcohol syndrome, are purely environmental in origin. Some of the syndromes that include periodontitis are caused by mutations in specific genes. For example, mutations in the cathepsin C gene have been shown to cause both Papillon–Lèfevre syndrome (Figure 6-2) and Haim–Munk syndrome as well as some forms of nonsyndromic prepubertal periodontitis, and they may also be associated with a risk of aggressive periodontitis.48 Periodontitis frequently occurs with some subtypes of Ehlers–Danlos syndrome, Kindler syndrome, Down syndrome (trisomy 21), leukocyte adhesion deficiencies (Figure 6-3), hypophosphatasia, two types of neutropenia, and aplasia of the lacrimal and salivary glands. A large triracial extended family demonstrated evidence of a single gene that caused both early-onset aggressive periodontitis and dentinogenesis imperfecta. The gene has been mapped with the use of linkage to a chromosomal region that contains a dentin matrix protein gene.37 Many of these conditions are so rare that few periodontists see even a single case during a lifetime of practice. However, dentists should be aware that these single-gene conditions exist; they need to be prepared to extend clinical evaluations to close relatives and to seek the assistance of or to refer to appropriately trained genetic counselors or specialists if a patient’s medical history or the presentation of multiple symptoms raises the possibility that he or she may be affected. Clinicians can obtain updated information about these conditions by accessing the publicly available Online Mendelian Inheritance in Man database and typing in “periodontitis OR periodontal disease” as the query term.2 Further research is necessary to determine whether inherited variation in the genes that cause these rare syndromes may also influence the risk of nonsyndromic forms of aggressive or chronic periodontitis.
Nonsyndromic Aggressive and Chronic Periodontitis
The reason why association studies of periodontitis have largely failed will be challenging to fully address. The first issue is straightforward and simply a matter of numbers. It is noteworthy that the successes achieved for many complex diseases (e.g., diabetes) with the use of the GWAS mapping approach were based on sample sizes involving thousands of cases and controls, with multiple replications by independent teams of investigators. Statistical theory shows that, to detect genes of modest effect, these large sample sizes are absolutely essential. With the use of a statistical power calculator developed for case–control studies,52 sample sizes required for 80% power are shown in Figure 6-4 for a study that involves only a single genetic marker as well as for studies that evaluate 5, 50, or 500 independent genetic markers in which the effects of multiple comparisons need to be accommodated. In this example, we assume that the risk gene acts in a dominant manner, with the high-risk allele occurring at a frequency of 25% in the population, and that this allele causes risk among carriers to increase by twofold (i.e., a greater effect on risk than observed for many susceptibility alleles found in GWAS studies of other complex diseases). Many periodontitis association studies reported in the literature involved multiple markers in each publication, and often the same research team reported positive findings for other genes in subsequent papers. Furthermore, because of the difficulties of publishing negative findings (i.e., when no association is found), many research teams working in this area may assay 50 or more genetic markers over the course of their work over several years. Results shown in Figure 6-4 demonstrate that, to obtain 80% power, a study of 50 markers would require more than 200 cases and 200 controls. Even if a research team assays only 5 SNPs, their study would still require 100 cases and 100 controls to achieve adequate power.
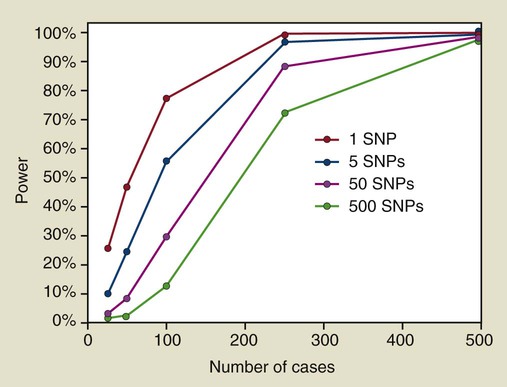
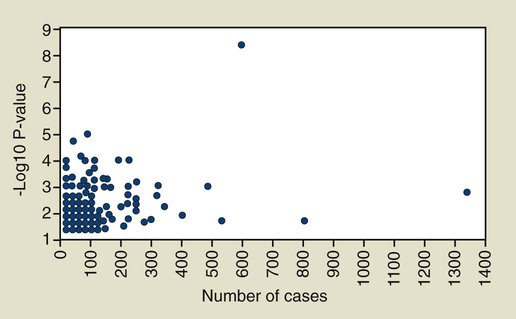
Our search of PubMed in early 2010 using the search term “(periodontal disease OR periodontitis) AND (SNP OR SNPs OR polymorphism OR polymorphisms OR linkage)” identified 311 periodontitis gene association tests with a P value of 0.05 or less for at least one statistical test reported. These results are summarized in Figure 6-5, in which the x-axis indicates the number of cases included in the study; the P value for the strongest finding is plotted on the y-axis. When more than one statistical test was reported, only the one with the smallest P value is shown in this figure. In some cases, this inflated the actual statistical significance, because investigators rarely adjust their findings for these multiple tests when the findings are reported in a publication. This analysis shows clearly that most association findings have been drastically underpowered if periodontitis is assumed to be a complex disease. The majority (66%) of these association reports for chronic and aggressive periodontitis are based on samples of 100 cases or less, and 41% are based on less than 60 cases. As shown in Figure 6-4, studies of such small sample size have little power to detect a susceptibility gene that increases risk by twofold. Given the added concern about publication bias (i.e., that positive findings are more likely to be accepted for publication), we can have little confidence that even the more statistically significant findings are valid and likely to be independently replicated if they are based on such very inadequate samples sizes. With only a few exceptions, which are noted later in this chapter, publications since 2010 have continued to involve small numbers of cases and to provide marginal statistical support for association.
Aside from important lessons about how not to carry out association studies of a complex disease, there are some tentative conclusions that can be drawn from the data available thus far. Detailed results of our review and manual curation of 298 publications reporting association findings for periodontitis are presented in Table 6-3, and key findings are condensed in Table 6-4.
TABLE 6-3
| Gene Symbol | Disease Type | Cases | Total | Best P Value | Level of Support | Population | Study Design | Stratified or Covariate Adj | Gene Name | Gene Aliases | # SNPs Assayed | SNP Name | PubMed ID | Journal |
| ABCA1 | CP | 22 | 41 | ns | None | Japanese | CC | N | ATP-binding cassette, sub-family A (ABC1), member 1 | TGD; ABC1; CERP; ABC-1; HDLDT1; FLJ14958; MGC164864; MGC165011; ABCA1 | 3 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ABCA4 | CP | 22 | 41 | ns | None | Japanese | CC | N | ATP-binding cassette, sub-family A (ABC1), member 4 | FFM; RMP; ABCR; RP19; STGD; ABC10; ARMD2; CORD3; STGD1; FLJ17534; DKFZp781N1972; ABCA4 | 2 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ABCB11 | CP | 22 | 41 | ns | None | Japanese | CC | N | ATP-binding cassette, sub-family B (MDR/TAP), member 11 | BSEP; PGY4; SPGP; ABC16; BRIC2; PFIC2; PFIC-2; ABCB11 | 7 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ABCC6 | CP | 22 | 41 | ns | None | Japanese | CC | N | ATP-binding cassette, sub-family C (CFTR/MRP), member 6 | ARA; PXE; MLP1; MRP6; PXE1; ABC34; MOATE; EST349056; ABCC6 | 9 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ABCC8 | CP | 22 | 41 | ns | None | Japanese | CC | N | ATP-binding cassette, sub-family C (CFTR/MRP), member 8 | HI; SUR; HHF1; MRP8; PHHI; SUR1; ABC36; HRINS; TNDM2; ABCC8 | 8 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ABP1 | CP | 22 | 41 | ns | None | Japanese | CC | N | Amiloride binding protein 1 (amine oxidase (copper-containing)) | ABP; AOC1; DAO; DAO1; KAO; ABP1 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ACACB | CP | 22 | 41 | ns | None | Japanese | CC | N | Acetyl-Coenzyme A (CoA) carboxylase beta | ACC2; ACCB; HACC275; ACACB | 2 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ACADL | CP | 22 | 41 | ns | None | Japanese | CC | N | Acyl-Coenzyme A dehydrogenase, long chain | LCAD; ACAD4; FLJ94052; ACADL | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ACAN | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Aggrecan | AGC1; SEDK; AGCAN; CSPG1; MSK16; CSPGCP; ACAN | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| ACE | AgP | 103 | 203 | ns | None | Turkish | CC | N | Angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 | DCP; ACE1; DCP1; CD143; MVCD3; MGC26566; ACE | 1 | I/D | 19162259 | Gürkan A, Emingil G, Saygan BH, Atilla G, Köse T, Baylas H, Berdeli A. Angiotensin-converting enzyme (ACE), angiotensinogen (AGT), and angiotensin II type 1 receptor (AT1R) gene polymorphisms in generalized aggressive periodontitis. Arch Oral Biol. 2009 Apr;54(4):337-44. Epub 2009 Jan 21. |
| ACE | CP | 90 | 216 | 0.015 | Weak | Turkish | CC | N | Angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 | DCP; ACE1; DCP1; CD143; MVCD3; MGC26566; ACE | 1 | I/D | 19236533 | Gürkan A, Emingil G, Saygan BH, Atilla G, Köse T, Baylas H, Berdeli A. Renin-angiotensin gene polymorphisms in relation to severe chronic periodontitis. J Clin Periodontol. 2009 Mar;36(3):204-11. |
| ACE | CP | 22 | 41 | ns | None | Japanese | CC | N | Angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 | DCP; ACE1; DCP1; CD143; MVCD3; MGC26566; ACE | 5 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ACE | CP | 63 | 158 | ns | None | Caucasian | CC | N | Angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 | DCP; ACE1; DCP1; CD143; MVCD3; MGC26566; ACE | 1 | Intron 16 I/D | 11210078 | Hollá LI, Fassmann A, Vasku A, Znojil V, Vanek J, Vácha J. Interactions of lymphotoxin alpha (TNF-beta), angiotensin-converting enzyme (ACE), and endothelin-1 (ET-1) gene polymorphisms in adult periodontitis. J Periodontol. 2001 Jan;72(1):85-9. |
| ACE.LTA | CP | 63 | 158 | 0.0228 | Weak | Caucasian | CC | N | n/a | n/a | 2 | Multiple Genes | 11210078 | Hollá LI, Fassmann A, Vasku A, Znojil V, Vanek J, Vácha J. Interactions of lymphotoxin alpha (TNF-beta), angiotensin-converting enzyme (ACE), and endothelin-1 (ET-1) gene polymorphisms in adult periodontitis. J Periodontol. 2001 Jan;72(1):85-9. |
| ACE2 | CP | 22 | 41 | ns | None | Japanese | CC | N | Angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 | ACEH; DKFZp434A014; ACE2 | 3 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ACSM3 | CP | 22 | 41 | ns | None | Japanese | CC | N | Acyl-CoA synthetase medium-chain family member 3 | SA; SAH; ACSM3 | 4 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ADA | CP | 22 | 41 | ns | None | Japanese | CC | N | Adenosine deaminase | ADA | 5 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ADAMTS8 | CP | 22 | 41 | ns | None | Japanese | CC | N | ADAM metallopeptidase with thrombospondin type 1 motif, 8 | METH2; ADAM-TS8; FLJ41712; ADAMTS8 | 2 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ADCY3 | CP | 22 | 41 | ns | None | Japanese | CC | N | Adenylate cyclase 3 | AC3; KIAA0511; ADCY3 | 3 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ADCY6 | CP | 22 | 41 | ns | None | Japanese | CC | N | Adenylate cyclase 6 | AC6; KIAA0422; DKFZp779F075; ADCY6 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ADCY9 | CP | 22 | 41 | ns | None | Japanese | CC | N | Adenylate cyclase 9 | AC9; ADCY9 | 7 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ADD1 | CP | 22 | 41 | ns | None | Japanese | CC | N | Adducin 1 (alpha) | ADDA; MGC3339; MGC44427; ADD1 | 6 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ADD2 | CP | 22 | 41 | ns | None | Japanese | CC | N | Adducin 2 (beta) | ADDB; ADD2 | 6 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ADH1C | CP | 22 | 41 | ns | None | Japanese | CC | N | Alcohol dehydrogenase 1C (class I), gamma polypeptide | ADH3; ADH1C | 5 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ADO22 | CP | 22 | 41 | ns | None | Japanese | CC | N | n/a | n/a | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ADORA2A | CP | 22 | 41 | ns | None | Japanese | CC | N | Adenosine A2a receptor | RDC8; hA2aR; ADORA2; ADORA2A | 3 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| AGER | CP | 101 | 263 | 0.02 | Weak | Caucasian | CC | N | Advanced glycosylation end product-specific receptor | RAGE; MGC22357; AGER | 3 | intron 7 (1704G/T) | 11811511 | Hollá LI, Kanková K, Fassmann A, Bucková D, Halabala T, Znojil V, Vanek J. Distribution of the receptor for advanced glycation end products gene polymorphisms in patients with chronic periodontitis: a preliminary study. J Periodontol. 2001 Dec;72(12):1742-6. |
| AGER | CP | 22 | 41 | ns | None | Japanese | CC | N | Advanced glycosylation end product-specific receptor | RAGE; MGC22357; AGER | 2 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| AGT | AgP | 103 | 203 | 0.004 | Weak | Turkish | CC | N | Angiotensinogen (serpin peptidase inhibitor, clade A, member 8) | ANHU; FLJ92595; FLJ97926; SERPINA8; AGT | 1 | M235T | 19162259 | Gürkan A, Emingil G, Saygan BH, Atilla G, Köse T, Baylas H, Berdeli A. Angiotensin-converting enzyme (ACE), angiotensinogen (AGT), and angiotensin II type 1 receptor (AT1R) gene polymorphisms in generalized aggressive periodontitis. Arch Oral Biol. 2009 Apr;54(4):337-44. Epub 2009 Jan 21. |
| AGT | CP | 90 | 216 | ns | None | Turkish | CC | N | Angiotensinogen (serpin peptidase inhibitor, clade A, member 8) | ANHU; FLJ92595; FLJ97926; SERPINA8; AGT | 1 | M235T | 19236533 | Gürkan A, Emingil G, Saygan BH, Atilla G, Köse T, Baylas H, Berdeli A. Renin-angiotensin gene polymorphisms in relation to severe chronic periodontitis. J Clin Periodontol. 2009 Mar;36(3):204-11. |
| AGTR1 | AgP | 103 | 203 | ns | None | Turkish | CC | N | Angiotensin II receptor, type 1 | AT1; AG2S; AT1B; AT1R; AT2R1; HAT1R; AGTR1A; AGTR1B; AT2R1A; AT2R1B; AGTR1 | 1 | A1166C | 19162259 | Gürkan A, Emingil G, Saygan BH, Atilla G, Köse T, Baylas H, Berdeli A. Angiotensin-converting enzyme (ACE), angiotensinogen (AGT), and angiotensin II type 1 receptor (AT1R) gene polymorphisms in generalized aggressive periodontitis. Arch Oral Biol. 2009 Apr;54(4):337-44. Epub 2009 Jan 21. |
| AGTR1 | CP | 90 | 216 | 0.03 | Weak | Turkish | CC | N | Angiotensin II receptor, type 1 | AT1; AG2S; AT1B; AT1R; AT2R1; HAT1R; AGTR1A; AGTR1B; AT2R1A; AT2R1B; AGTR1 | 1 | A1166C | 19236533 | Gürkan A, Emingil G, Saygan BH, Atilla G, Köse T, Baylas H, Berdeli A. Renin-angiotensin gene polymorphisms in relation to severe chronic periodontitis. J Clin Periodontol. 2009 Mar;36(3):204-11. |
| AIRE | CP | 22 | 41 | ns | None | Japanese | CC | N | Autoimmune regulator | APS1; APSI; PGA1; AIRE1; APECED; AIRE | 6 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ALCAM | CP | 22 | 41 | ns | None | Japanese | CC | N | Activated leukocyte cell adhesion molecule | MEMD; CD166; FLJ38514; MGC71733; ALCAM | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ALDH2 | CP | 22 | 41 | ns | None | Japanese | CC | N | Aldehyde dehydrogenase 2 family (mitochondriAl) | ALDM; ALDHI; ALDH-E2; MGC1806; ALDH2 | 3 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ALDH3A1 | CP | 22 | 41 | ns | None | Japanese | CC | N | Aldehyde dehydrogenase 3 family, member A1 | ALDH3; ALDHIII; MGC10406; ALDH3A1 | 3 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ALDH3A2 | CP | 22 | 41 | ns | None | Japanese | CC | N | Aldehyde dehydrogenase 3 family, member A2 | SLS; FALDH; ALDH10; FLJ20851; DKFZp686E23276; ALDH3A2 | 3 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ALOX15 | CP | 22 | 41 | ns | None | Japanese | CC | N | Arachidonate 15-lipoxygenase | 15LOX-1; 15-LOX-1; ALOX15 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| AMACR | CP | 22 | 41 | ns | None | Japanese | CC | N | Alpha-methylacyl-CoA racemase | RACE; CBAS4; AMACR | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| AMPD1 | CP | 22 | 41 | ns | None | Japanese | CC | N | Adenosine monophosphate deaminase 1 (isoform M) | MAD; MADA; AMPD1 | 3 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| APOA4 | CP | 22 | 41 | ns | None | Japanese | CC | N | Apolipoprotein A-IV | MGC142154; MGC142156; APOA4 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| APOH | CP | 22 | 41 | ns | None | Japanese | CC | N | Apolipoprotein H (beta-2-glycoprotein I) | BG; B2G1; APOH | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| APOL1 | CP | 22 | 41 | ns | None | Japanese | CC | N | Apolipoprotein L, 1 | APOL; APO-L; APOL-I; APOL1 | 4 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| AQP8 | CP | 22 | 41 | ns | None | Japanese | CC | N | Aquaporin 8 | AQP8 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ART1 | CP | 22 | 41 | ns | None | Japanese | CC | N | ADP-ribosyltransferase 1 | RT6; ART2; CD296; MGC133217; ART1 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ASIP | CP | 22 | 41 | ns | None | Japanese | CC | N | Agouti signaling protein, nonagouti homolog (mouse) | ASP; AGSW; AGTI; AGTIL; SHEP9; MGC126092; MGC126093; ASIP | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| B3GNT3 | CP | 22 | 41 | ns | None | Japanese | CC | N | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 | TMEM3; B3GN-T3; B3GNT-3; HP10328; B3GAL-T8; beta3Gn-T3; B3GNT3 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| BAT1 | CP&HIV | 16 | 32 | 0.0002 | Weak | Caucasian | CC | G | HLA-B associated transcript 1 | UAP56; D6S81E; DDX39B; BAT1 | 1 | HLA-B8 | 10551423 | Price P, Calder DM, Witt CS, Allcock RJ, Christiansen FT, Davies GR, Cameron PU, Rogers M, Baluchova K, Moore CB, French MA. Periodontal attachment loss in HIV-infected patients is associated with the Major histocompatibility complex 8.1 haplotype (HLA-A1,B8,DR3). Tissue Antigens. 1999 Oct;54(4):391-9. |
| BDKRB1 | CP | 22 | 41 | ns | None | Japanese | CC | N | Bradykinin receptor B1 | B1R; BKR1; B1BKR; BKB1R; BRADYB1; BDKRB1 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| BDKRB2 | CP | 22 | 41 | ns | None | Japanese | CC | N | Bradykinin receptor B2 | B2R; BK2; BK-2; BKR2; BRB2; DKFZp686O088; BDKRB2 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| BMP2 | AgP | 98 | 186 | ns | None | Japanese | CC | N | Bone morphogenetic protein 2 | BMP2A; BMP2 | 1 | rs1049007 | 16844084 | Rabello D, Soedarsono N, Kamei H, Ishihara Y, Noguchi T, Fuma D, Suzuki M, Sakaki Y, Yamaguchi A, Kojima T. CSF1 gene associated with aggressive periodontitis in the Japanese population. Biochem Biophys Res Commun. 2006 Sep 1;347(3):791-6. Epub 2006 Jul 7. |
| BMP3 | AgP | 98 | 186 | ns | None | Japanese | CC | N | Bone morphogenetic protein 3 | BMP3A; BMP3 | 1 | rs3733549 | 16844084 | Rabello D, Soedarsono N, Kamei H, Ishihara Y, Noguchi T, Fuma D, Suzuki M, Sakaki Y, Yamaguchi A, Kojima T. CSF1 gene associated with aggressive periodontitis in the Japanese population. Biochem Biophys Res Commun. 2006 Sep 1;347(3):791-6. Epub 2006 Jul 7. |
| BMP4 | AgP | 98 | 186 | ns | None | Japanese | CC | N | Bone morphogenetic protein 4 | ZYME; BMP2B; OFC11; BMP2B1; MCOPS6; BMP4 | 1 | rs2071047 | 16844084 | Rabello D, Soedarsono N, Kamei H, Ishihara Y, Noguchi T, Fuma D, Suzuki M, Sakaki Y, Yamaguchi A, Kojima T. CSF1 gene associated with aggressive periodontitis in the Japanese population. Biochem Biophys Res Commun. 2006 Sep 1;347(3):791-6. Epub 2006 Jul 7. |
| BMP7 | AgP | 98 | 186 | ns | None | Japanese | CC | N | Bone morphogenetic protein 7 | OP-1; BMP7 | 6 | Multiple SNPs | 16844084 | Rabello D, Soedarsono N, Kamei H, Ishihara Y, Noguchi T, Fuma D, Suzuki M, Sakaki Y, Yamaguchi A, Kojima T. CSF1 gene associated with aggressive periodontitis in the Japanese population. Biochem Biophys Res Commun. 2006 Sep 1;347(3):791-6. Epub 2006 Jul 7. |
| BMP8B | CP | 22 | 41 | ns | None | Japanese | CC | N | Bone morphogenetic protein 8b | OP2; BMP8; MGC131757; BMP8B | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| BPI | CP | 123 | 255 | ns | None | German | CC | N | Bactericidal/permeability-increasing protein | BPI | 1 | Lys216Glu | 16893388 | Glas J, Török HP, Tonenchi L, Hamann S, Malachova O, Euba A, Folwaczny C, Folwaczny M. A645G (Lys216Glu) polymorphism of the bactericidal/permeability-increasing protein gene in periodontal disease. Int J Immunogenet. 2006 Aug;33(4):255-60. |
| C5 | AgP+CP | 229 | 436 | 0.001 | Weak | Chinese | CC | AGS | Complement Component 5 | CPAMD4; FLJ17816; FLJ17822; MGC142298; C5 | 4 | Haplotype | 19909405 | Chai L, Song YQ, Zee KY, Leung WK. Single nucleotide polymorphisms of complement component 5 and periodontitis. J Periodontal Res. 2009 Nov 9. |
| C5 | AgP+CP | 229 | 436 | 0.007 | Weak | Chinese | CC | AGS | Complement Component 5 | CPAMD4; FLJ17816; FLJ17822; MGC142298; C5 | 11 | rs17611 | 19909405 | Chai L, Song YQ, Zee KY, Leung WK. Single nucleotide polymorphisms of complement component 5 and periodontitis. J Periodontal Res. 2009 Nov 9. |
| C9 | CP | 22 | 41 | ns | None | Japanese | CC | N | Complement Component 9 | C9 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| CALCR | CP | 22 | 41 | 0.0039 | Weak | Japanese | CC | N | Calcitonin receptor | CRT; CTR; CTR1; CALCR | 7 | IMS-JST054515 | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| CASR | CP | 22 | 41 | ns | None | Japanese | CC | N | Calcium-sensing receptor | CAR; FHH; FIH; HHC; EIG8; HHC1; NSHPT; PCAR1; GPRC2A; MGC138441; CASR | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| CAT | CP | 22 | 41 | ns | None | Japanese | CC | N | Catalase | MGC138422; MGC138424; CAT | 5 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| CCK | CP | 22 | 41 | ns | None | Japanese | CC | N | Cholecystokinin | MGC117187; CCK | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| CCKAR | CP | 22 | 41 | ns | None | Japanese | CC | N | Cholecystokinin A receptor | CCK-A; CCKRA; CCK1-R; CCKAR | 2 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| CCL15 | CP | 22 | 41 | ns | None | Japanese | CC | N | Chemokine (C-C motif) ligand 15 | LKN1; NCC3; SY15; HCC-2; Lkn-1; MIP-5; NCC-3; SCYL3; MIP-1d; SCYA15; HMRP-2B; CCL15 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| CCL25 | CP | 22 | 41 | ns | None | Japanese | CC | N | Chemokine (C-C motif) ligand 25 | TECK; Ckb15; SCYA25; MGC150327; CCL25 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| CCL5 | CP | 106 | 175 | ns | None | Caucasian | CC | S | Chemokine (C-C motif) ligand 5 | SISd; SCYA5; RANTES; TCP228; D17S136E; MGC17164; CCL5 | 1 | −471 A/G | 17305874 | Savarrio L, Donati M, Carr C, Kinane DF, Berglundh T. Interleukin-24, RANTES and CCR5 gene polymorphisms are not associated with chronic adult periodontitis. J Periodontal Res. 2007 Apr;42(2):152-8. |
| CCR2 | CP | 22 | 41 | ns | None | Japanese | CC | N | Chemokine (C-C motif) receptor 2 | CKR2; CCR2A; CCR2B; CD192; CKR2A; CKR2B; CMKBR2; MCP-1-R; CC-CKR-2; FLJ78302; MGC103828; MGC111760; MGC168006; CCR2 | 2 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| CCR5 | CP | 106 | 175 | ns | None | Caucasian | CC | S | Chemokine (C-C motif) receptor 5 | CKR5; CD195; CKR-5; CCCKR5; CMKBR5; IDDM22; CC-CKR-5; FLJ78003; CCR5 | 1 | Delta32/wt | 17305874 | Savarrio L, Donati M, Carr C, Kinane DF, Berglundh T. Interleukin-24, RANTES and CCR5 gene polymorphisms are not associated with chronic adult periodontitis. J Periodontal Res. 2007 Apr;42(2):152-8. |
| CCR5 | CP | 137 | 219 | ns | None | Caucasian | CC | GS | Chemokine (C-C motif) receptor 5 | CKR5; CD195; CKR-5; CCCKR5; CMKBR5; IDDM22; CC-CKR-5; FLJ78003; CCR5 | 1 | 59653 C>T | 16512757 | Wohlfahrt JC, Wu T, Hodges JS, Hinrichs JE, Michalowicz BS. No association between selected candidate gene polymorphisms and severe chronic periodontitis. J Periodontol. 2006 Mar;77(3):426-36. |
| CCR5 | CP | 22 | 41 | ns | None | Japanese | CC | N | Chemokine (C-C motif) receptor 5 | CKR5; CD195; CKR-5; CCCKR5; CMKBR5; IDDM22; CC-CKR-5; FLJ78003; CCR5 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| CCR5 | CP | 81 | 202 | ns | None | Caucasian | CC | N | Chemokine (C-C motif) receptor 5 | CKR5; CD195; CKR-5; CCCKR5; CMKBR5; IDDM22; CC-CKR-5; FLJ78003; CCR5 | 1 | Delta32/wt | 14697747 | Folwaczny M, Glas J, Török HP, Fricke K, Folwaczny C. Prevalence of the chemokine receptor CCR5-Delta32 gene mutation in periodontal disease. Clin Immunol. 2003 Dec;109(3):325-9. |
| CD14 | AgP | 73 | 153 | ns | None | German | CC | N | CD14 molecule | CD14 | 1 | −159 | 19046305 | Schulz S, Zissler N, Altermann W, Klapproth J, Zimmermann U, Gläser C, Schaller HG, Reichert S. Impact of genetic variants of CD14 and TLR4 on subgingival periodontopathogens. Int J Immunogenet. 2008 Dec;35(6):457-64. |
| CD14 | AgP | 73 | 200 | ns | None | Caucasian | CC | N | CD14 molecule | CD14 | 2 | Multiple SNPs | 17309585 | James JA, Poulton KV, Haworth SE, Payne D, McKay IJ, Clarke FM, Hughes FJ, Linden GJ. Polymorphisms of TLR4 but not CD14 are associated with a decreased risk of aggressive periodontitis. J Clin Periodontol. 2007 Feb;34(2):111-7. |
| CD14 | AgP+CP | 133 | 213 | ns | None | German | CC | N | CD14 molecule | CD14 | 1 | −159 | 19046305 | Schulz S, Zissler N, Altermann W, Klapproth J, Zimmermann U, Gläser C, Schaller HG, Reichert S. Impact of genetic variants of CD14 and TLR4 on subgingival periodontopathogens. Int J Immunogenet. 2008 Dec;35(6):457-64. |
| CD14 | AgP+CP | 319 | 622 | ns | None | Japanese | CC | AGS | CD14 molecule | CD14 | 1 | −159 | 19892918 | Kobayashi T, Nagata T, Murakami S, Takashiba S, Kurihara H, Izumi Y, Numabe Y, Watanabe H, Kataoka M, Nagai A, Hayashi J, Ohyama H, Okamatsu Y, Inagaki Y, Tai H, Yoshie H. Genetic risk factors for periodontitis in a Japanese population. J Dent Res. 2009 Dec;88(12):1137-41. |
| CD14 | CP | 100 | 199 | 0.004 | Weak | Caucasian | CC | AGPS | CD14 molecule | CD14 | 1 | −260 | 16246938 | Laine ML, Morré SA, Murillo LS, van Winkelhoff AJ, Peña AS. CD14 and TLR4 gene polymorphisms in adult periodontitis. J Dent Res. 2005 Nov;84(11):1042-6. |
| CD14 | CP | 98 | 134 | 0.006 | Weak | Caucasian | CC | N | CD14 molecule | CD14 | 2 | G-1359T | 12414826 | Holla LI, Buckova D, Fassmann A, Halabala T, Vasku A, Vacha J. Promoter polymorphisms in the CD14 receptor gene and their potential association with the severity of chronic periodontitis. J Med Genet. 2002 Nov;39(11):844-8. |
| CD14 | CP | 70 | 145 | 0.013 | Weak | German | CC | G | CD14 molecule | CD14 | 1 | −159 | 15491315 | Folwaczny M, Glas J, Török HP, Fricke K, Folwaczny C. The CD14 -159C-to-T promoter polymorphism in periodontal disease. J Clin Periodontol. 2004 Nov;31(11):991-5. |
| CD14 | CP | 51 | 229 | 0.022 | Weak | Caucasian | CC | GS | CD14 molecule | CD14 | 1 | −260 | 17448042 | Tervonen T, Raunio T, Knuuttila M, Karttunen R. Polymorphisms in the CD14 and IL-6 genes associated with periodontal disease. J Clin Periodontol. 2007 May;34(5):377-83. |
| CD14 | CP | 60 | 99 | 0.028 | Weak | Caucasian | CC | N | CD14 molecule | CD14 | 1 | −159 | 15842262 | Donati M, Berglundh T, Hytönen AM, Hahn-Zoric M, Hanson LA, Padyukov L. Association of the -159 CD14 gene polymorphism and lack of association of the -308 TNFA and Q551R IL-4RA polymorphisms with severe chronic periodontitis in Swedish Caucasians. J Clin Periodontol. 2005 May;32(5):474-9. |
| CD14 | CP | 56 | 84 | ns | None | Finnish | CC | N | CD14 molecule | CD14 | 1 | −260 | 19500269 | Raunio T, Knuuttila M, Karttunen R, Vainio O, Tervonen T. Serum sCD14, polymorphism of CD14(-260) and periodontal infection. Oral Dis. 2009 Jun 4. [Epub ahead of print] |
| CD14 | CP | 105 | 162 | ns | None | Caucasian | CC | N | CD14 molecule | CD14 | 1 | −260 | 19318422 | Nicu EA, Laine ML, Morré SA, Van der Velden U, Loos BG. Soluble CD14 in periodontitis. Innate Immun. 2009 Apr;15(2):121-8. |
| CD14 | CP | 60 | 140 | ns | None | German | CC | N | CD14 molecule | CD14 | 1 | −159 | 19046305 | Schulz S, Zissler N, Altermann W, Klapproth J, Zimmermann U, Gläser C, Schaller HG, Reichert S. Impact of genetic variants of CD14 and TLR4 on subgingival periodontopathogens. Int J Immunogenet. 2008 Dec;35(6):457-64. |
| CD14 | CP | n/a | 80 | ns | None | Finnish | Quan | N | CD14 molecule | CD14 | 1 | −260 | 19017034 | Raunio T, Knuuttila M, Hiltunen L, Karttunen R, Vainio O, Tervonen T. IL-6(-174) genotype associated with the extent of periodontal disease in type 1 diabetic subjects. J Clin Periodontol. 2009 Jan;36(1):11-7. Epub 2008 Oct 31. |
| CD14 | CP | 95 | 189 | ns | None | Caucasian | CC | N | CD14 molecule | CD14 | 2 | Multiple SNPs | 17309585 | James JA, Poulton KV, Haworth SE, Payne D, McKay IJ, Clarke FM, Hughes FJ, Linden GJ. Polymorphisms of TLR4 but not CD14 are associated with a decreased risk of aggressive periodontitis. J Clin Periodontol. 2007 Feb;34(2):111-7. |
| CD14 | CP | 163 | 267 | ns | None | Japanese | CC | N | CD14 molecule | CD14 | 1 | −159 | 12885845 | Yamazaki K, Ueki-Maruyama K, Oda T, Tabeta K, Shimada Y, Tai H, Nakajima T, Yoshie H, Herawati D, Seymour GJ. Single-nucleotide polymorphism in the CD14 promoter and periodontal disease expression in a Japanese population. J Dent Res. 2003 Aug;82(8):612-6. |
| CD14.IL6 | CP | 51 | 229 | 0.012 | Weak | Caucasian | CC | GS | n/a | n/a | 2 | Multiple Genes | 17448042 | Tervonen T, Raunio T, Knuuttila M, Karttunen R. Polymorphisms in the CD14 and IL-6 genes associated with periodontal disease. J Clin Periodontol. 2007 May;34(5):377-83. |
| CD14.TLR4 | CP | 51 | 229 | ns | None | Caucasian | CC | GS | n/a | n/a | 2 | Multiple Genes | 17448042 | Tervonen T, Raunio T, Knuuttila M, Karttunen R. Polymorphisms in the CD14 and IL-6 genes associated with periodontal disease. J Clin Periodontol. 2007 May;34(5):377-83. |
| CD2 | CP | 22 | 41 | ns | None | Japanese | CC | N | CD2 molecule | T11; SRBC; FLJ46032; CD2 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| CD36 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | CD36 molecule (thrombospondin receptor) | FAT; GP4; GP3B; GPIV; CHDS7; PASIV; SCARB3; CD36 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| CD36 | CP | 22 | 41 | ns | None | Japanese | CC | N | CD36 molecule (thrombospondin receptor) | FAT; GP4; GP3B; GPIV; CHDS7; PASIV; SCARB3; CD36 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| CD4 | CP | 22 | 41 | ns | None | Japanese | CC | N | CD4 molecule | CD4mut; CD4 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| CD6 | CP | 22 | 41 | ns | None | Japanese | CC | N | CD6 molecule | TP120; FLJ44171; CD6 | 4 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| CD81 | CP | 22 | 41 | ns | None | Japanese | CC | N | CD81 molecule | S5.7; TAPA1; TSPAN28; CD81 | 3 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| CDKN2BAS | AgP | 159 | 895 | 0.00069 | Moderate | German | CC | DGS | CDKN2B antisense RNA (non-protein coding) | ANRIL; p15AS; NCRNA00089; RP11-145E5.4; CDKN2BAS | 3 | rs1333048 | 19214202 | Schaefer AS, Richter GM, Groessner-Schreiber B, Noack B, Nothnagel M, El Mokhtari NE, Loos BG, Jepsen S, Schreiber S. Identification of a shared genetic susceptibility locus for coronary heart disease and periodontitis. PLoS Genet. 2009 Feb;5(2):e1000378. Epub 2009 Feb 13. |
| CDKN2BAS | AgP | 146 | 514 | 0.0059 | Weak | German | CC | DGS | CDKN2B antisense RNA (non-protein coding) | ANRIL; p15AS; NCRNA00089; RP11-145E5.4; CDKN2BAS | 3 | rs1333048 | 19214202 | Schaefer AS, Richter GM, Groessner-Schreiber B, Noack B, Nothnagel M, El Mokhtari NE, Loos BG, Jepsen S, Schreiber S. Identification of a shared genetic susceptibility locus for coronary heart disease and periodontitis. PLoS Genet. 2009 Feb;5(2):e1000378. Epub 2009 Feb 13. |
| CHGB | CP | 22 | 41 | ns | None | Japanese | CC | N | Chromogranin B (secretogranin 1) | SCG1; CHGB | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| CHI3L1 | CP | 22 | 41 | ns | None | Japanese | CC | N | Chitinase 3-like 1 (cartilage glycoprotein-39) | GP39; ASRT7; YKL40; YYL-40; HC-gp39; HCGP-3P; FLJ38139; DKFZp686N19119; CHI3L1 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| Chr19 Region | CP | 300 | 600 | 0.019 | Weak | Japanese | CC | N | n/a | n/a | 13 | rs11668269 | 19335087 | Tabeta K, Shimada Y, Tai H, Ishihara Y, Noguchi T, Soga Y, Takashiba S, Suzuki G, Kobayashi T, Oka A, Kobayashi T, Yamazaki K, Inoko H, Yoshie H. Assessment of chromosome 19 for genetic association in severe chronic periodontitis. J Periodontol. 2009 Apr;80(4):663-71. |
| CKAP2L | AgP | 415 | 1289 | ns | None | Caucasian | CC | N | Cytoskeleton associated protein 2-like | FLJ40629; MGC39683; CKAP2L | 2 | Multiple SNPs | 18723088 | Fiebig A, Jepsen S, Loos BG, Scholz C, Schäfer C, Rühling A, Nothnagel M, Eickholz P, van der Velden U, Schenck K, Schreiber S, Grössner-Schreiber B. Polymorphisms in the interleukin-1 (IL1) gene cluster are not associated with aggressive periodontitis in a large Caucasian population. Genomics. 2008 Nov;92(5):309-15. Epub 2008 Sep 18. |
| CKAP2L | AgP | 415 | 1289 | ns | None | Caucasian | CC | N | Cytoskeleton associated protein 2-like | FLJ40629; MGC39683; CKAP2L | 2 | Haplotype | 18723088 | Fiebig A, Jepsen S, Loos BG, Scholz C, Schäfer C, Rühling A, Nothnagel M, Eickholz P, van der Velden U, Schenck K, Schreiber S, Grössner-Schreiber B. Polymorphisms in the interleukin-1 (IL1) gene cluster are not associated with aggressive periodontitis in a large Caucasian population. Genomics. 2008 Nov;92(5):309-15. Epub 2008 Sep 18. |
| COL12A1 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Collagen, type XII, alpha 1 | COL12A1L; BA209D8.1; DJ234P15.1; COL12A1 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| COL15A1 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Collagen, type XV, alpha 1 | FLJ38566; COL15A1 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| COL16A1 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Collagen, type XVI, alpha 1 | 447AA; FP1572; COL16A1 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| COL17A1 | AgP+CP | 251 | 376 | 0.008 | Weak | Japanese | CC | N | Collagen, type XVII, alpha 1 | BP180; BPAG2; LAD-1; FLJ60881; KIAA0204; BA16H23.2; COL17A1 | n/a | Chr:10 C/T | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| COL18A1 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Collagen, type XVIII, alpha 1 | KNO; KNO1; FLJ27325; FLJ34914; MGC74745; COL18A1 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| COL19A1 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Collagen, type XIX, alpha 1 | COL9A1L; D6S228E; COL19A1 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| COL1A1 | AgP | 46 | 136 | ns | None | Greek | CC | N | Collagen, type I, alpha 1 | OI4; COL1A1 | 1 | Sp1 | 16911569 | Sakellari D, Katsares V, Georgiadou M, Kouvatsi A, Arsenakis M, Konstantinidis A. No correlation of five gene polymorphisms with periodontal conditions in a Greek population. J Clin Periodontol. 2006 Nov;33(11):765-70. Epub 2006 Aug 14. |
| COL1A1 | AgP+CP | 251 | 376 | 0.003 | Weak | Japanese | CC | N | Collagen, type I, alpha 1 | OI4; COL1A1 | n/a | Chr:17 A/C | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| COL1A1 | CP | 56 | 146 | ns | None | Greek | CC | N | Collagen, type I, alpha 1 | OI4; COL1A1 | 1 | Sp1 | 16911569 | Sakellari D, Katsares V, Georgiadou M, Kouvatsi A, Arsenakis M, Konstantinidis A. No correlation of five gene polymorphisms with periodontal conditions in a Greek population. J Clin Periodontol. 2006 Nov;33(11):765-70. Epub 2006 Aug 14. |
| COL1A2 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Collagen, type I, alpha 2 | OI4; COL1A2 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| COL2A1 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Collagen, type II, alpha 1 | AOM; ANFH; SEDC; COL11A3; MGC131516; COL2A1 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| COL3A1 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Collagen, type III, alpha 1 | EDS4A; FLJ34534; COL3A1 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| COL4A1 | AgP+CP | 251 | 376 | 0.003 | Weak | Japanese | CC | N | Collagen, type IV, alpha 1 | arresten; COL4A1 | n/a | Chr:13 T/C | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| COL4A3 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Collagen, type IV, alpha 3 (Goodpasture antigen) | COL4A3 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| COL4A4 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Collagen, type IV, alpha 4 | CA44; COL4A4 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| COL5A3 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Collagen, type V, alpha 3 | COL5A3 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| COL6A2 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Collagen, type VI, alpha 2 | PP3610; FLJ46862; DKFZp586E1322; COL6A2 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| COL6A3 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Collagen, type VI, alpha 3 | FLJ34702; FLJ98399; DKFZp686N0262; DKFZp686D23123; DKFZp686K04147; COL6A3 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| COL7A1 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Collagen, type VII, alpha 1 | EBD1; EBR1; EBDCT; COL7A1 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| COL8A1 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Collagen, type VIII, alpha 1 | C3orf7; MGC9568; COL8A1 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| COL9A1 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Collagen, type IX, alpha 1 | MED; EDM6; FLJ40263; DJ149L1.1.2; COL9A1 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| COL9A2 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Collagen, type IX, alpha 2 | MED; EDM2; DJ39G22.4; COL9A2 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| COLQ | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase | EAD; FLJ55041; COLQ | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| CSF1 | AgP | 98 | 186 | 0.024698 | Weak | Japanese | CC | N | Colony stimulating factor 1 (macrophage) | MCSF; MGC31930; CSF1 | 5 | G/T | 16844084 | Rabello D, Soedarsono N, Kamei H, Ishihara Y, Noguchi T, Fuma D, Suzuki M, Sakaki Y, Yamaguchi A, Kojima T. CSF1 gene associated with aggressive periodontitis in the Japanese population. Biochem Biophys Res Commun. 2006 Sep 1;347(3):791-6. Epub 2006 Jul 7. |
| CST1 | CP | 22 | 41 | ns | None | Japanese | CC | N | Cystatin SN | CST1 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| CST5 | CP | 22 | 41 | ns | None | Japanese | CC | N | Cystatin D | MGC71922; CST5 | 5 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| CTF2 | CP | 22 | 41 | ns | None | Japanese | CC | N | n/a | n/a | 3 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| CTLA4 | CP | 137 | 219 | ns | None | Caucasian | CC | GS | Cytotoxic T-lymphoCyte-associated protein 4 | CD; GSE; ICOS; CD152; CTLA-4; IDDM12; CELIAC3; CTLA4 | 1 | 49 A>G | 16512757 | Wohlfahrt JC, Wu T, Hodges JS, Hinrichs JE, Michalowicz BS. No association between selected candidate gene polymorphisms and severe chronic periodontitis. J Periodontol. 2006 Mar;77(3):426-36. |
| CTSB | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Cathepsin B | APPS; CPSB; CTSB | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| CTSB | CP | 22 | 41 | ns | None | Japanese | CC | N | Cathepsin B | APPS; CPSB; CTSB | 4 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| CTSC | AgP | 110 | 188 | 0.034 | Weak | German | CC | N | Cathepsin C | JP; HMS; JPD; PLS; CPPI; DPP1; DPPI; PALS; CTSC | 5 | p.I453V | 18809751 | Noack B, Görgens H, Hempel U, Fanghänel J, Hoffmann T, Ziegler A, Schackert HK. Cathepsin C gene variants in aggressive periodontitis. J Dent Res. 2008 Oct;87(10):958-63. |
| CTSC | PPP | 3 | 7 | ns | None | n/a | Fam | N | Cathepsin C | JP; HMS; JPD; PLS; CPPI; DPP1; DPPI; PALS; CTSC | 3 | Multiple SNPs | 14974080 | Hewitt C, McCormick D, Linden G, Turk D, Stern I, Wallace I, Southern L, Zhang L, Howard R, Bullon P, Wong M, Widmer R, Gaffar KA, Awawdeh L, Briggs J, Yaghmai R, Jabs EW, Hoeger P, Bleck O, Rüdiger SG, Petersilka G, Battino M, Brett P, Hattab F, Al-Hamed M, Sloan P, Toomes C, Dixon M, James J, Read AP, Thakker N. The role of cathepsin C in Papillon-Lefèvre syndrome, prepubertal periodontitis, and aggressive periodontitis. Hum Mutat. 2004 Mar;23(3):222-8. |
| CTSC | PPP | 4 | 60 | sig | Weak | Jordanian | Fam | N | Cathepsin C | JP; HMS; JPD; PLS; CPPI; DPP1; DPPI; PALS; CTSC | 1 | 1040A->G | 10662808 | Hart TC, Hart PS, Michalec MD, Zhang Y, Marazita ML, Cooper M, Yassin OM, Nusier M, Walker S. Localisation of a gene for prepubertal periodontitis to chromosome 11q14 and identification of a cathepsin C gene mutation. J Med Genet. 2000 Feb;37(2):95-101. |
| CTSC | PPP | 14 | 14 | sig | Weak | Jordanian | Fam | N | Cathepsin C | JP; HMS; JPD; PLS; CPPI; DPP1; DPPI; PALS; CTSC | 1 | 1040A-G | 10662808 | Hart TC, Hart PS, Michalec MD, Zhang Y, Marazita ML, Cooper M, Yassin OM, Nusier M, Walker S. Localisation of a gene for prepubertal periodontitis to chromosome 11q14 and identification of a cathepsin C gene mutation. J Med Genet. 2000 Feb;37(2):95-101. |
| CTSD | AgP+CP | 251 | 376 | 0.0007 | Moderate | Japanese | CC | N | Cathepsin D | CPSD; CLN10; MGC2311; CTSD | n/a | Chr:11 C/T | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| CTSD | CP | 22 | 41 | ns | None | Japanese | CC | N | Cathepsin D | CPSD; CLN10; MGC2311; CTSD | 3 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| CTSG | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Cathepsin G | CG; MGC23078; CTSG | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| CTSH | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Cathepsin H | CPSB; ACC-4; ACC-5; MGC1519; minichain; DKFZp686B24257; CTSH | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| CTSL1 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Cathepsin L1 | MEP; CATL; CTSL; FLJ31037; CTSL1 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| CYBA | AgP | 224 | 455 | 0.002 | Weak | Mixed | CC | EGS | Cytochrome b-245, alpha polypeptide | p22-PHOX; CYBA | 1 | C242T | 16899095 | Nibali L, Parkar M, Brett P, Knight J, Tonetti MS, Griffiths GS. NADPH oxidase (CYBA) and FcgammaR polymorphisms as risk factors for aggressive periodontitis: a case-control association study. J Clin Periodontol. 2006 Aug;33(8):529-39. |
| CYBA | CP | 22 | 41 | ns | None | Japanese | CC | N | Cytochrome b-245, alpha polypeptide | p22-PHOX; CYBA | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| CYBA.FCGR3B | AgP | 224 | 455 | 0.001 | Weak | Mixed | CC | EGS | n/a | n/a | 2 | Multiple Genes | 16899095 | Nibali L, Parkar M, Brett P, Knight J, Tonetti MS, Griffiths GS. NADPH oxidase (CYBA) and FcgammaR polymorphisms as risk factors for aggressive periodontitis: a case-control association study. J Clin Periodontol. 2006 Aug;33(8):529-39. |
| CYP1A1 | CP | 115 | 241 | 0.009 | Weak | Korean | CC | S | Cytochrome P450, family 1, subfamily A, polypeptide 1 | AHH; AHRR; CP11; CYP1; P1-450; P450-C; P450DX; CYP1A1 | 1 | Msp1 | 15491310 | Kim JS, Park JY, Chung WY, Choi MA, Cho KS, Park KK. Polymorphisms in genes coding for enzymes metabolizing smoking-derived substances and the risk of periodontitis. J Clin Periodontol. 2004 Nov;31(11):959-64. |
| CYP21A2 | CP | 22 | 41 | ns | None | Japanese | CC | N | Cytochrome P450, family 21, subfamily A, polypeptide 2 | CAH1; CPS1; CA21H; CYP21; CYP21B; P450c21B; MGC150536; MGC150537; CYP21A2 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| CYP2E1 | CP | 115 | 241 | 0.05 | Weak | Korean | CC | S | Cytochrome P450, family 2, subfamily E, polypeptide 1 | CPE1; CYP2E; P450-J; P450C2E; CYP2E1 | 1 | Pst1 | 15491310 | Kim JS, Park JY, Chung WY, Choi MA, Cho KS, Park KK. Polymorphisms in genes coding for enzymes metabolizing smoking-derived substances and the risk of periodontitis. J Clin Periodontol. 2004 Nov;31(11):959-64. |
| CYP4B1 | CP | 22 | 41 | ns | None | Japanese | CC | N | Cytochrome P450, family 4, subfamily B, polypeptide 1 | CYPIVB1; P-450HP; CYP4B1 | 2 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| CYP4F2 | CP | 22 | 41 | ns | None | Japanese | CC | N | Cytochrome P450, family 4, subfamily F, polypeptide 2 | CPF2; CYP4F2 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| DEFB1 | AgP | 532 | 2004 | 0.02 | Weak | Caucasian | CC | SD | Defensin, beta 1 | D1; HBD1; DEFB-1; DEFB101; MGC51822; DEFB1 | 12 | Multiple SNPs | 19829306 | Schaefer AS, Richter GM, Nothnagel M, Laine ML, Rühling A, Schäfer C, Cordes N, Noack B, Folwaczny M, Glas J, Dörfer C, Dommisch H, Groessner-Schreiber B, Jepsen S, Loos BG, Schreiber S. A 3’ UTR transition within DEFB1 is associated with chronic and aggressive periodontitis. Genes Immun. 2009 Oct 15. [Epub ahead of print] |
| DEFB1 | AgP | 120 | 280 | ns | None | Mixed | CC | N | Defensin, beta 1 | D1; HBD1; DEFB-1; DEFB101; MGC51822; DEFB1 | 1 | −44 | 15242954 | Boniotto M, Hazbón MH, Jordan WJ, Lennon GP, Eskdale J, Alland D, Gallagher G. Novel hairpin-shaped primer assay to study the association of the -44 single-nucleotide polymorphism of the DEFB1 gene with early-onset periodontal disease. Clin Diagn Lab Immunol. 2004 Jul;11(4):766-9. |
| DEFB1 | AgP+CP | 1337 | 3128 | 0.002 | Weak | Caucasian | CC | SD | Defensin, beta 1 | D1; HBD1; DEFB-1; DEFB101; MGC51822; DEFB1 | 12 | Multiple SNPs | 19829306 | Schaefer AS, Richter GM, Nothnagel M, Laine ML, Rühling A, Schäfer C, Cordes N, Noack B, Folwaczny M, Glas J, Dörfer C, Dommisch H, Groessner-Schreiber B, Jepsen S, Loos BG, Schreiber S. A 3’ UTR transition within DEFB1 is associated with chronic and aggressive periodontitis. Genes Immun. 2009 Oct 15. [Epub ahead of print] |
| DEFB1 | CP | 805 | 1124 | 0.02 | Weak | Caucasian | CC | SD | Defensin, beta 1 | D1; HBD1; DEFB-1; DEFB101; MGC51822; DEFB1 | 12 | Multiple SNPs | 19829306 | Schaefer AS, Richter GM, Nothnagel M, Laine ML, Rühling A, Schäfer C, Cordes N, Noack B, Folwaczny M, Glas J, Dörfer C, Dommisch H, Groessner-Schreiber B, Jepsen S, Loos BG, Schreiber S. A 3’ UTR transition within DEFB1 is associated with chronic and aggressive periodontitis. Genes Immun. 2009 Oct 15. [Epub ahead of print] |
| DEFB1 | CP | 137 | 219 | ns | None | Caucasian | CC | GS | Defensin, beta 1 | D1; HBD1; DEFB-1; DEFB101; MGC51822; DEFB1 | 1 | 692 G>A | 16512757 | Wohlfahrt JC, Wu T, Hodges JS, Hinrichs JE, Michalowicz BS. No association between selected candidate gene polymorphisms and severe chronic periodontitis. J Periodontol. 2006 Mar;77(3):426-36. |
| DEFB1 | CP | 22 | 41 | ns | None | Japanese | CC | N | Defensin, beta 1 | D1; HBD1; DEFB-1; DEFB101; MGC51822; DEFB1 | 3 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| DPP4 | CP | 22 | 41 | 0.006 | Weak | Japanese | CC | N | Dipeptidyl-peptidase 4 | CD26; ADABP; ADCP2; DPPIV; TP103; DPP4 | 7 | IMS-JST022880 | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ECE1 | CP | 22 | 41 | ns | None | Japanese | CC | N | Endothelin converting enzyme 1 | ECE; ECE1 | 7 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ECH1 | CP | 22 | 41 | ns | None | Japanese | CC | N | Enoyl Coenzyme A hydratase 1, peroxisomal | HPXEL; ECH1 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| EDN1 | CP | 22 | 41 | ns | None | Japanese | CC | N | Endothelin 1 | ET1; HDLCQ7; EDN1 | 4 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| EDN1 | CP | 63 | 158 | ns | None | Caucasian | CC | N | Endothelin 1 | ET1; HDLCQ7; EDN1 | 1 | Intron4 TaqI | 11210078 | Hollá LI, Fassmann A, Vasku A, Znojil V, Vanek J, Vácha J. Interactions of lymphotoxin alpha (TNF-beta), angiotensin-converting enzyme (ACE), and endothelin-1 (ET-1) gene polymorphisms in adult periodontitis. J Periodontol. 2001 Jan;72(1):85-9. |
| EDN1.LTA | CP | 63 | 158 | 0.0472 | Weak | Caucasian | CC | N | n/a | n/a | 2 | Multiple Genes | 11210078 | Hollá LI, Fassmann A, Vasku A, Znojil V, Vanek J, Vácha J. Interactions of lymphotoxin alpha (TNF-beta), angiotensin-converting enzyme (ACE), and endothelin-1 (ET-1) gene polymorphisms in adult periodontitis. J Periodontol. 2001 Jan;72(1):85-9. |
| EDN3 | CP | 22 | 41 | ns | None | Japanese | CC | N | Endothelin 3 | ET3; MGC15067; MGC61498; EDN3 | 4 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| EGF | AgP+CP | 251 | 376 | 0.004 | Weak | Japanese | CC | N | Epidermal growth factor (beta-urogastrone) | URG; HOMG4; EGF | n/a | Chr:4 A/G | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| EGF | CP | 22 | 41 | ns | None | Japanese | CC | N | Epidermal growth factor (beta-urogastrone) | URG; HOMG4; EGF | 10 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| EGFR | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | ERBB; HER1; mENA; ERBB1; PIG61; EGFR | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| EIF2AK2 | CP | 22 | 41 | ns | None | Japanese | CC | N | Eukaryotic translation initiation factor 2-alpha kinase 2 | PKR; PRKR; EIF2AK1; MGC126524; EIF2AK2 | 2 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ELN | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Elastin | WS; WBS; SVAS; FLJ38671; FLJ43523; ELN | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| EMR1 | CP | 22 | 41 | ns | None | Japanese | CC | N | EGF-like module containing, mucin-like, hormone receptor-like 1 | TM7LN3; EMR1 | 4 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ENTPD6 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Ectonucleoside triphosphate diphosphohydrolase 6 (putative function) | CD39L2; IL6ST2; IL-6SAG; FLJ36711; NTPDase-6; dJ738P15.3; DKFZp781G2277; DKFZp781K21102; ENTPD6 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| EPHX1 | CP | 22 | 41 | ns | None | Japanese | CC | N | Epoxide hydrolase 1, microsomal (xenobiotic) | MEH; EPHX; EPOX; EPHX1 | 2 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| EPYC | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Epiphycan | PGLB; DSPG3; Pg-Lb; SLRR3B; EPYC | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| ERAP1 | CP | 22 | 41 | ns | None | Japanese | CC | N | Endoplasmic reticulum aminopeptidase 1 | ALAP; A-LAP; ARTS1; ERAAP; APPILS; ARTS-1; ERAAP1; PILSAP; PILS-AP; KIAA0525; ERAP1 | 4 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ESR1 | AgP | 90 | 181 | ns | None | Chinese | CC | G | Estrogen receptor 1 | ER; ESR; Era; ESRA; NR3A1; DKFZp686N23123; ESR1 | 2 | Multiple SNPs | 15324358 | Zhang L, Meng H, Zhao H, Li Q, Xu L, Chen Z, Shi D, Feng X. Estrogen receptor-alpha gene polymorphisms in patients with periodontitis. J Periodontal Res. 2004 Oct;39(5):362-6. |
| ESR1 | AgP | n/a | n/a | sig | Weak | Chinese | CC | N | Estrogen receptor 1 | ER; ESR; Era; ESRA; NR3A1; DKFZp686N23123; ESR1 | 1 | n/a | 19031816 | Wang HY, Pan YP. [Screening and analysis of multi-alleles in generalized aggressive periodontitis]. Zhonghua Kou Qiang Yi Xue Za Zhi. 2008 Jul;43(7):406-9. Chinese. |
| ESR1 | CP | 34 | 125 | <0.01 | Weak | Chinese | CC | G | Estrogen receptor 1 | ER; ESR; Era; ESRA; NR3A1; DKFZp686N23123; ESR1 | 2 | XbaI | 15324358 | Zhang L, Meng H, Zhao H, Li Q, Xu L, Chen Z, Shi D, Feng X. Estrogen receptor-alpha gene polymorphisms in patients with periodontitis. J Periodontal Res. 2004 Oct;39(5):362-6. |
| ESR1 | CP | n/a | n/a | sig | Weak | Chinese | CC | N | Estrogen receptor 1 | ER; ESR; Era; ESRA; NR3A1; DKFZp686N23123; ESR1 | 2 | XbaI | 18846948 | Wang HY, Pan YP, Teng D, Zhao J, Lin L. [The relativity between chronic periodontitis and the genetic polymorphisms of vitamin D receptor and estrogen receptor]. Zhonghua Kou Qiang Yi Xue Za Zhi. 2008 Apr;43(4):236-9. Chinese. |
| ESR2 | CP | 22 | 41 | ns | None | Japanese | CC | N | Estrogen receptor 2 (ER beta) | Erb; ESRB; ESTRB; NR3A2; ER-BETA; ESR-BETA; ESR2 | 5 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| ETFA | CP | 22 | 41 | ns | None | Japanese | CC | N | Electron-transfer-flavoprotein, alpha polypeptide | EMA; GA2; MADD; ETFA | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| F5 | CP | 22 | 41 | ns | None | Japanese | CC | N | Coagulation factor V (proaccelerin, labile factor) | FVL; PCCF; F5 | 6 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| FABP2 | CP | 22 | 41 | ns | None | Japanese | CC | N | Fatty acid binding protein 2, intestinal | FABPI; I-FABP; MGC133132; FABP2 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| FABP7 | CP | 22 | 41 | ns | None | Japanese | CC | N | Fatty acid binding protein 7, brain | MRG; BLBP; FABPB; B-FABP; DKFZp547J2313; FABP7 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| FAS | CP | 22 | 41 | ns | None | Japanese | CC | N | Fas (TNF receptor superfamily, member 6) | APT1; CD95; FAS1; APO-1; FASTM; ALPS1A; TNFRSF6; FAS | 3 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| FASLG | CP | 137 | 219 | ns | None | Caucasian | CC | GS | Fas ligand (TNF superfamily, member 6) | FASL; CD178; CD95L; TNFSF6; APT1LG1; FASLG | 1 | −844 C>T | 16512757 | Wohlfahrt JC, Wu T, Hodges JS, Hinrichs JE, Michalowicz BS. No association between selected candidate gene polymorphisms and severe chronic periodontitis. J Periodontol. 2006 Mar;77(3):426-36. |
| FASLG | CP | 22 | 41 | ns | None | Japanese | CC | N | Fas ligand (TNF superfamily, member 6) | FASL; CD178; CD95L; TNFSF6; APT1LG1; FASLG | 3 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| FBP1 | CP | 22 | 41 | ns | None | Japanese | CC | N | Fructose-1,6-bisphosphatase 1 | FBP; FBP1 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| FCAR | AgP | 46 | 126 | 0.014 | Weak | Japanese | CC | N | Fc fragment of IgA, receptor for | CD89; FCAR | 1 | nt 324 A/G | 15140034 | Kaneko S, Kobayashi T, Yamamoto K, Jansen MD, van de Winkel JG, Yoshie H. A novel polymorphism of FcalphaRI (CD89) associated with aggressive periodontitis. Tissue Antigens. 2004 Jun;63(6):572-7. |
| FCAR | AgP | 46 | 126 | 0.014 | Weak | Japanese | CC | N | Fc fragment of IgA, receptor for | CD89; FCAR | 1 | nt 324 A/G | 15140034 | Kaneko S, Kobayashi T, Yamamoto K, Jansen MD, van de Winkel JG, Yoshie H. A novel polymorphism of FcalphaRI (CD89) associated with aggressive periodontitis. Tissue Antigens. 2004 Jun;63(6):572-7. |
| FCAR | AgP | 224 | 455 | ns | None | Mixed | CC | EGS | Fc fragment of IgA, receptor for | CD89; FCAR | 1 | 324 A-G | 16899095 | Nibali L, Parkar M, Brett P, Knight J, Tonetti MS, Griffiths GS. NADPH oxidase (CYBA) and FcgammaR polymorphisms as risk factors for aggressive periodontitis: a case-control association study. J Clin Periodontol. 2006 Aug;33(8):529-39. |
| FCAR | AgP+CP | 319 | 622 | ns | None | Japanese | CC | AGS | Fc fragment of IgA, receptor for | CD89; FCAR | 2 | n/a | 19892918 | Kobayashi T, Nagata T, Murakami S, Takashiba S, Kurihara H, Izumi Y, Numabe Y, Watanabe H, Kataoka M, Nagai A, Hayashi J, Ohyama H, Okamatsu Y, Inagaki Y, Tai H, Yoshie H. Genetic risk factors for periodontitis in a Japanese population. J Dent Res. 2009 Dec;88(12):1137-41. |
| FCAR | CP | 113 | 221 | ns | None | Japanese | CC | N | Fc fragment of IgA, receptor for | CD89; FCAR | 2 | Multiple SNPs | 18321309 | Komatsu Y, Galicia JC, Kobayashi T, Yamazaki K, Yoshie H. Association of interleukin-1 receptor antagonist +2018 gene polymorphism with Japanese chronic periodontitis patients using a novel genotyping method. Int J Immunogenet. 2008 Apr;35(2):165-70. |
| FCAR.TGFB1 | CP | 113 | 221 | ns | None | Japanese | CC | N | n/a | n/a | 3 | Multiple Genes | 18321309 | Komatsu Y, Galicia JC, Kobayashi T, Yamazaki K, Yoshie H. Association of interleukin-1 receptor antagonist +2018 gene polymorphism with Japanese chronic periodontitis patients using a novel genotyping method. Int J Immunogenet. 2008 Apr;35(2):165-70. |
| FCGR2A | AgP | 12 | 73 | 0.013 | Weak | Caucasian | CC | S | Fc fragment of IgG, low affinity IIa, receptor (CD32) | CD32; FCG2; FcGR; CD32A; CDw32; FCGR2; IGFR2; FCGR2A1; MGC23887; MGC30032; FCGR2A | 1 | +131 | 12834496 | Loos BG, Leppers-Van de Straat FG, Van de Winkel JG, Van der Velden U. Fcgamma receptor polymorphisms in relation to periodontitis. J Clin Periodontol. 2003 Jul;30(7):595-602. |
| FCGR2A | AgP | 20 | 72 | ns | None | Polish | CC | N | Fc fragment of IgG, low affinity IIa, receptor (CD32) | CD32; FCG2; FcGR; CD32A; CDw32; FCGR2; IGFR2; FCGR2A1; MGC23887; MGC30032; FCGR2A | 1 | CD32 | 19127818 | Drozdzik A. [The effect of environmental and genetic factors in the pathogenesis of periodontitis]. Ann Acad Med Stetin. 2008;54(1):118-26. Polish. |
| FCGR2A | AgP | 224 | 455 | ns | None | Mixed | CC | EGS | Fc fragment of IgG, low affinity IIa, receptor (CD32) | CD32; FCG2; FcGR; CD32A; CDw32; FCGR2; IGFR2; FCGR2A1; MGC23887; MGC30032; FCGR2A | 1 | 494 A/G | 16899095 | Nibali L, Parkar M, Brett P, Knight J, Tonetti MS, Griffiths GS. NADPH oxidase (CYBA) and FcgammaR polymorphisms as risk factors for aggressive periodontitis: a case-control association study. J Clin Periodontol. 2006 Aug;33(8):529-39. |
| FCGR2A | AgP | 31 | 80 | ns | None | Brazilian | CC | EG | Fc fragment of IgG, low affinity IIa, receptor (CD32) | CD32; FCG2; FcGR; CD32A; CDw32; FCGR2; IGFR2; FCGR2A1; MGC23887; MGC30032; FCGR2A | 1 | +131 | 16805673 | de Souza RC, Colombo AP. Distribution of FcgammaRIIa and FcgammaRIIIb genotypes in patients with generalized aggressive periodontitis. J Periodontol. 2006 Jul;77(7):1120-8. |
| FCGR2A | AgP | 32 | 104 | ns | None | Japanese | CC | N | Fc fragment of IgG, low affinity IIa, receptor (CD32) | CD32; FCG2; FcGR; CD32A; CDw32; FCGR2; IGFR2; FCGR2A1; MGC23887; MGC30032; FCGR2A | 1 | n/a | 14647193 | Yasuda K, Sugita N, Kobayashi T, Yamamoto K, Yoshie H. FcgammaRIIB gene polymorphisms in Japanese periodontitis patients. Genes Immun. 2003 Dec;4(8):541-6. |
| FCGR2A | AgP | 33 | 60 | ns | None | Chinese | CC | N | Fc fragment of IgG, low affinity IIa, receptor (CD32) | CD32; FCG2; FcGR; CD32A; CDw32; FCGR2; IGFR2; FCGR2A1; MGC23887; MGC30032; FCGR2A | 1 | n/a | 11776881 | Fu Y, Cao C, Wang S. [Relevance of Fc gamma R polymorphism to the susceptibility of early-onset periodontitis]. Zhonghua Kou Qiang Yi Xue Za Zhi. 1999 Nov;34(6):364-6. Chinese. |
| FCGR2A | AgP | 38 | 142 | ns | None | Japanese | CC | N | Fc fragment of IgG, low affinity IIa, receptor (CD32) | CD32; FCG2; FcGR; CD32A; CDw32; FCGR2; IGFR2; FCGR2A1; MGC23887; MGC30032; FCGR2A | 1 | +131 | 11022771 | Kobayashi T, Sugita N, van der Pol WL, Nunokawa Y, Westerdaal NA, Yamamoto K, van de Winkel JG, Yoshie H. The Fcgamma receptor genotype as a risk factor for generalized early-onset periodontitis in Japanese patients. J Periodontol. 2000 Sep;71(9):1425-32. |
| FCGR2A | AgP | 48 | 115 | ns | None | African American | CC | N | Fc fragment of IgG, low affinity IIa, receptor (CD32) | CD32; FCG2; FcGR; CD32A; CDw32; FCGR2; IGFR2; FCGR2A1; MGC23887; MGC30032; FCGR2A | n/a | 131H/R | 12027254 | Fu Y, Korostoff JM, Fine DH, Wilson ME. Fc gamma receptor genes as risk markers for localized aggressive periodontitis in African-Americans. J Periodontol. 2002 May;73(5):517-23. |
| FCGR2A | AgP+CP | 319 | 622 | ns | None | Japanese | CC | AGS | Fc fragment of IgG, low affinity IIa, receptor (CD32) | CD32; FCG2; FcGR; CD32A; CDw32; FCGR2; IGFR2; FCGR2A1; MGC23887; MGC30032; FCGR2A | 1 | R131/H131 | 19892918 | Kobayashi T, Nagata T, Murakami S, Takashiba S, Kurihara H, Izumi Y, Numabe Y, Watanabe H, Kataoka M, Nagai A, Hayashi J, Ohyama H, Okamatsu Y, Inagaki Y, Tai H, Yoshie H. Genetic risk factors for periodontitis in a Japanese population. J Dent Res. 2009 Dec;88(12):1137-41. |
| FCGR2A | CP | 213 | 422 | 0.03 | Weak | Caucasian | CC | AGS | Fc fragment of IgG, low affinity IIa, receptor (CD32) | CD32; FCG2; FcGR; CD32A; CDw32; FCGR2; IGFR2; FCGR2A1; MGC23887; MGC30032; FCGR2A | 1 | +131 | 15152814 | Yamamoto K, Kobayashi T, Grossi S, Ho AW, Genco RJ, Yoshie H, De Nardin E. Association of Fcgamma receptor IIa genotype with chronic periodontitis in Caucasians. J Periodontol. 2004 Apr;75(4):517-22. |
| FCGR2A | CP | 166 | 246 | <0.0125 | Weak | Chinese | CC | N | Fc fragment of IgG, low affinity IIa, receptor (CD32) | CD32; FCG2; FcGR; CD32A; CDw32; FCGR2; IGFR2; FCGR2A1; MGC23887; MGC30032; FCGR2A | 1 | +131 | 15190804 | Tang Y, Zhang JC, Zhang WH, Pang RY. [The association between Fc gamma receptor IIA gene polymorphism and susceptibility to chronic periodontitis in Chinese Han nationality]. Hua Xi Kou Qiang Yi Xue Za Zhi. 2004 Apr;22(2):158-61. Chinese. |
| FCGR2A | CP | 132 | 205 | <0.05 | Weak | Caucasian | CC | N | Fc fragment of IgG, low affinity IIa, receptor (CD32) | CD32; FCG2; FcGR; CD32A; CDw32; FCGR2; IGFR2; FCGR2A1; MGC23887; MGC30032; FCGR2A | 1 | +131 | 16889631 | Wolf DL, Neiderud AM, Hinckley K, Dahlén G, van de Winkel JG, Papapanou PN. Fcgamma receptor polymorphisms and periodontal status: a prospective follow-up study. J Clin Periodontol. 2006 Oct;33(10):691-8. Epub 2006 Aug 3. |
| FCGR2A | CP | 32 | 84 | ns | None | Polish | CC | N | Fc fragment of IgG, low affinity IIa, receptor (CD32) | CD32; FCG2; FcGR; CD32A; CDw32; FCGR2; IGFR2; FCGR2A1; MGC23887; MGC30032; FCGR2A | 1 | CD32 | 19127818 | Drozdzik A. [The effect of environmental and genetic factors in the pathogenesis of periodontitis]. Ann Acad Med Stetin. 2008;54(1):118-26. Polish. |
| FCGR2A | CP | 113 | 221 | ns | None | Japanese | CC | N | Fc fragment of IgG, low affinity IIa, receptor (CD32) | CD32; FCG2; FcGR; CD32A; CDw32; FCGR2; IGFR2; FCGR2A1; MGC23887; MGC30032; FCGR2A | 1 | +131 | 18321309 | Komatsu Y, Galicia JC, Kobayashi T, Yamazaki K, Yoshie H. Association of interleukin-1 receptor antagonist +2018 gene polymorphism with Japanese chronic periodontitis patients using a novel genotyping method. Int J Immunogenet. 2008 Apr;35(2):165-70. |
| FCGR2A | CP | 100 | 200 | ns | None | Japanese | CC | N | Fc fragment of IgG, low affinity IIa, receptor (CD32) | CD32; FCG2; FcGR; CD32A; CDw32; FCGR2; IGFR2; FCGR2A1; MGC23887; MGC30032; FCGR2A | 1 | nt 559 G/T | 18052703 | Kobayashi T, Ito S, Kuroda T, Yamamoto K, Sugita N, Narita I, Sumida T, Gejyo F, Yoshie H. The interleukin-1 and Fcgamma receptor gene polymorphisms in Japanese patients with rheumatoid arthritis and periodontitis. J Periodontol. 2007 Dec;78(12):2311-8. |
| FCGR2A | CP | 50 | 124 | ns | None | Taiwanese | CC | N | Fc fragment of IgG, low affinity IIa, receptor (CD32) | CD32; FCG2; FcGR; CD32A; CDw32; FCGR2; IGFR2; FCGR2A1; MGC23887; MGC30032; FCGR2A | 1 | +131 | 14761117 | Chung HY, Lu HC, Chen WL, Lu CT, Yang YH, Tsai CC. Gm (23) allotypes and Fcgamma receptor genotypes as risk factors for various forms of periodontitis. J Clin Periodontol. 2003 Nov;30(11):954-60. |
| FCGR2A | CP | 72 | 144 | ns | None | Japanese | CC | N | Fc fragment of IgG, low affinity IIa, receptor (CD32) | CD32; FCG2; FcGR; CD32A; CDw32; FCGR2; IGFR2; FCGR2A1; MGC23887; MGC30032; FCGR2A | 1 | n/a | 14647193 | Yasuda K, Sugita N, Kobayashi T, Yamamoto K, Yoshie H. FcgammaRIIB gene polymorphisms in Japanese periodontitis patients. Genes Immun. 2003 Dec;4(8):541-6. |
| FCGR2A | CP | 56 | 117 | ns | None | Caucasian | CC | S | Fc fragment of IgG, low affinity IIa, receptor (CD32) | CD32; FCG2; FcGR; CD32A; CDw32; FCGR2; IGFR2; FCGR2A1; MGC23887; MGC30032; FCGR2A | 1 | +131 | 12834496 | Loos BG, Leppers-Van de Straat FG, Van de Winkel JG, Van der Velden U. Fcgamma receptor polymorphisms in relation to periodontitis. J Clin Periodontol. 2003 Jul;30(7):595-602. |
| FCGR2A | CP | 89 | 153 | ns | None | Japanese | CC | S | Fc fragment of IgG, low affinity IIa, receptor (CD32) | CD32; FCG2; FcGR; CD32A; CDw32; FCGR2; IGFR2; FCGR2A1; MGC23887; MGC30032; FCGR2A | 1 | +131 | 11699473 | Kobayashi T, Yamamoto K, Sugita N, van der Pol WL, Yasuda K, Kaneko S, van de Winkel JG, Yoshie H. The Fc gamma receptor genotype as a severity factor for chronic periodontitis in Japanese patients. J Periodontol. 2001 Oct;72(10):1324-31. |
| FCGR2A | CP | 49 | 98 | ns | None | German | CC | AS | Fc fragment of IgG, low affinity IIa, receptor (CD32) | CD32; FCG2; FcGR; CD32A; CDw32; FCGR2; IGFR2; FCGR2A1; MGC23887; MGC30032; FCGR2A | 1 | +131 | 11528518 | Meisel P, Carlsson LE, Sawaf H, Fanghaenel J, Greinacher A, Kocher T. Polymorphisms of Fc gamma-receptors RIIa, RIIIa, and RIIIb in patients with adult periodontal diseases. Genes Immun. 2001 Aug;2(5):258-62. |
| FCGR2A | CP | 83 | 187 | ns | None | Japanese | CC | N | Fc fragment of IgG, low affinity IIa, receptor (CD32) | CD32; FCG2; FcGR; CD32A; CDw32; FCGR2; IGFR2; FCGR2A1; MGC23887; MGC30032; FCGR2A | 1 | +131 | 11022771 | Kobayashi T, Sugita N, van der Pol WL, Nunokawa Y, Westerdaal NA, Yamamoto K, van de Winkel JG, Yoshie H. The Fcgamma receptor genotype as a risk factor for generalized early-onset periodontitis in Japanese patients. J Periodontol. 2000 Sep;71(9):1425-32. |
| FCGR2A | CP | 100 | 205 | ns | None | Japanese | CC | N | Fc fragment of IgG, low affinity IIa, receptor (CD32) | CD32; FCG2; FcGR; CD32A; CDw32; FCGR2; IGFR2; FCGR2A1; MGC23887; MGC30032; FCGR2A | 1 | +131 | 9284119 | Kobayashi T, Westerdaal NA, Miyazaki A, van der Pol WL, Suzuki T, Yoshie H, van de Winkel JG, Hara K. Relevance of immunoglobulin G Fc receptor polymorphism to recurrence of adult periodontitis in Japanese patients. Infect Immun. 1997 Sep;65(9):3556-60. |
| FCGR2A | CP | 86 | 113 | ns | None | n/a | CC | S | Fc fragment of IgG, low affinity IIa, receptor (CD32) | CD32; FCG2; FcGR; CD32A; CDw32; FCGR2; IGFR2; FCGR2A1; MGC23887; MGC30032; FCGR2A | 1 | R131H | 9667480 | Colombo AP, Eftimiadi C, Haffajee AD, Cugini MA, Socransky SS. Serum IgG2 level, Gm(23) allotype and FcgammaRIIa and FcgammaRIIIb receptors in refractory periodontal disease. J Clin Periodontol. 1998 Jun;25(6):465-74. |
| FCGR2A | CP&RA | 86 | 186 | ns | None | Japanese | CC | N | Fc fragment of IgG, low affinity IIa, receptor (CD32) | CD32; FCG2; FcGR; CD32A; CDw32; FCGR2; IGFR2; FCGR2A1; MGC23887; MGC30032; FCGR2A | 1 | nt 559 G/T | 18052703 | Kobayashi T, Ito S, Kuroda T, Yamamoto K, Sugita N, Narita I, Sumida T, Gejyo F, Yoshie H. The interleukin-1 and Fcgamma receptor gene polymorphisms in Japanese patients with rheumatoid arthritis and periodontitis. J Periodontol. 2007 Dec;78(12):2311-8. |
| FCGR2A | CP&SLE | 42 | 84 | 0.0013 | Weak | Japanese | CC | N | Fc fragment of IgG, low affinity IIa, receptor (CD32) | CD32; FCG2; FcGR; CD32A; CDw32; FCGR2; IGFR2; FCGR2A1; MGC23887; MGC30032; FCGR2A | 1 | +131 | 12710759 | Kobayashi T, Ito S, Yamamoto K, Hasegawa H, Sugita N, Kuroda T, Kaneko S, Narita I, Yasuda K, Nakano M, Gejyo F, Yoshie H. Risk of periodontitis in systemic lupus erythematosus is associated with Fcgamma receptor polymorphisms. J Periodontol. 2003 Mar;74(3):378-84. |
| FCGR2A | CP&SLE | 42 | 60 | 0.011 | Weak | Japanese | CC | N | Fc fragment of IgG, low affinity IIa, receptor (CD32) | CD32; FCG2; FcGR; CD32A; CDw32; FCGR2; IGFR2; FCGR2A1; MGC23887; MGC30032; FCGR2A | 1 | +131 | 12710759 | Kobayashi T, Ito S, Yamamoto K, Hasegawa H, Sugita N, Kuroda T, Kaneko S, Narita I, Yasuda K, Nakano M, Gejyo F, Yoshie H. Risk of periodontitis in systemic lupus erythematosus is associated with Fcgamma receptor polymorphisms. J Periodontol. 2003 Mar;74(3):378-84. |
| FCGR2A.FCGR3A | CP | 100 | 200 | ns | None | Japanese | CC | N | n/a | n/a | 2 | Multiple Genes | 18052703 | Kobayashi T, Ito S, Kuroda T, Yamamoto K, Sugita N, Narita I, Sumida T, Gejyo F, Yoshie H. The interleukin-1 and Fcgamma receptor gene polymorphisms in Japanese patients with rheumatoid arthritis and periodontitis. J Periodontol. 2007 Dec;78(12):2311-8. |
| FCGR2A.FCGR3A | CP&RA | 86 | 186 | ns | None | Japanese | CC | N | n/a | n/a | 2 | Multiple Genes | 18052703 | Kobayashi T, Ito S, Kuroda T, Yamamoto K, Sugita N, Narita I, Sumida T, Gejyo F, Yoshie H. The interleukin-1 and Fcgamma receptor gene polymorphisms in Japanese patients with rheumatoid arthritis and periodontitis. J Periodontol. 2007 Dec;78(12):2311-8. |
| FCGR2A.FCGR3B | AgP | 38 | 142 | 0.04 | Weak | Japanese | CC | N | n/a | n/a | 2 | Multiple Genes | 11022771 | Kobayashi T, Sugita N, van der Pol WL, Nunokawa Y, Westerdaal NA, Yamamoto K, van de Winkel JG, Yoshie H. The Fcgamma receptor genotype as a risk factor for generalized early-onset periodontitis in Japanese patients. J Periodontol. 2000 Sep;71(9):1425-32. |
| FCGR2A.FCGR3B | AgP | 31 | 80 | <0.001 | Weak | Brazilian | CC | EG | n/a | n/a | 2 | Multiple Genes | 16805673 | de Souza RC, Colombo AP. Distribution of FcgammaRIIa and FcgammaRIIIb genotypes in patients with generalized aggressive periodontitis. J Periodontol. 2006 Jul;77(7):1120-8. |
| FCGR2A.FCGR3B | CP | 100 | 200 | ns | None | Japanese | CC | N | n/a | n/a | 2 | Multiple Genes | 18052703 | Kobayashi T, Ito S, Kuroda T, Yamamoto K, Sugita N, Narita I, Sumida T, Gejyo F, Yoshie H. The interleukin-1 and Fcgamma receptor gene polymorphisms in Japanese patients with rheumatoid arthritis and periodontitis. J Periodontol. 2007 Dec;78(12):2311-8. |
| FCGR2A.FCGR3B | CP | 83 | 187 | ns | None | Japanese | CC | N | n/a | n/a | 2 | Multiple Genes | 11022771 | Kobayashi T, Sugita N, van der Pol WL, Nunokawa Y, Westerdaal NA, Yamamoto K, van de Winkel JG, Yoshie H. The Fcgamma receptor genotype as a risk factor for generalized early-onset periodontitis in Japanese patients. J Periodontol. 2000 Sep;71(9):1425-32. |
| FCGR2A.FCGR3B | CP&RA | 86 | 186 | ns | None | Japanese | CC | N | n/a | n/a | 2 | Multiple Genes | 18052703 | Kobayashi T, Ito S, Kuroda T, Yamamoto K, Sugita N, Narita I, Sumida T, Gejyo F, Yoshie H. The interleukin-1 and Fcgamma receptor gene polymorphisms in Japanese patients with rheumatoid arthritis and periodontitis. J Periodontol. 2007 Dec;78(12):2311-8. |
| FCGR2A.FCGR3B.FCGR2B | CP | 113 | 221 | ns | None | Japanese | CC | N | n/a | n/a | 3 | Multiple Genes | 18321309 | Komatsu Y, Galicia JC, Kobayashi T, Yamazaki K, Yoshie H. Association of interleukin-1 receptor antagonist +2018 gene polymorphism with Japanese chronic periodontitis patients using a novel genotyping method. Int J Immunogenet. 2008 Apr;35(2):165-70. |
| FCGR2B | AgP | 32 | 104 | 0.002 | Weak | Japanese | CC | N | Fc fragment of IgG, low affinity IIb, receptor (CD32) | CD32; FCG2; CD32B; FCGR2; IGFR2; FCGR2B | 11 | nt 695 (T-C) | 14647193 | Yasuda K, Sugita N, Kobayashi T, Yamamoto K, Yoshie H. FcgammaRIIB gene polymorphisms in Japanese periodontitis patients. Genes Immun. 2003 Dec;4(8):541-6. |
| FCGR2B | AgP | 224 | 455 | ns | None | Mixed | CC | EGS | Fc fragment of IgG, low affinity IIb, receptor (CD32) | CD32; FCG2; CD32B; FCGR2; IGFR2; FCGR2B | 1 | 695 T-C | 16899095 | Nibali L, Parkar M, Brett P, Knight J, Tonetti MS, Griffiths GS. NADPH oxidase (CYBA) and FcgammaR polymorphisms as risk factors for aggressive periodontitis: a case-control association study. J Clin Periodontol. 2006 Aug;33(8):529-39. |
| FCGR2B | AgP+CP | 319 | 622 | ns | None | Japanese | CC | AGS | Fc fragment of IgG, low affinity IIb, receptor (CD32) | CD32; FCG2; CD32B; FCGR2; IGFR2; FCGR2B | 2 | n/a | 19892918 | Kobayashi T, Nagata T, Murakami S, Takashiba S, Kurihara H, Izumi Y, Numabe Y, Watanabe H, Kataoka M, Nagai A, Hayashi J, Ohyama H, Okamatsu Y, Inagaki Y, Tai H, Yoshie H. Genetic risk factors for periodontitis in a Japanese population. J Dent Res. 2009 Dec;88(12):1137-41. |
| FCGR2B | CP | 72 | 144 | 0.011 | Weak | Japanese | CC | N | Fc fragment of IgG, low affinity IIb, receptor (CD32) | CD32; FCG2; CD32B; FCGR2; IGFR2; FCGR2B | 11 | nt 646 184 (A-G) | 14647193 | Yasuda K, Sugita N, Kobayashi T, Yamamoto K, Yoshie H. FcgammaRIIB gene polymorphisms in Japanese periodontitis patients. Genes Immun. 2003 Dec;4(8):541-6. |
| FCGR2B | CP | 113 | 221 | ns | None | Japanese | CC | N | Fc fragment of IgG, low affinity IIb, receptor (CD32) | CD32; FCG2; CD32B; FCGR2; IGFR2; FCGR2B | 1 | −232 | 18321309 | Komatsu Y, Galicia JC, Kobayashi T, Yamazaki K, Yoshie H. Association of interleukin-1 receptor antagonist +2018 gene polymorphism with Japanese chronic periodontitis patients using a novel genotyping method. Int J Immunogenet. 2008 Apr;35(2):165-70. |
| FCGR3A | AgP | 12 | 73 | 0.044 | Weak | Caucasian | CC | S | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | CD16; FCG3; CD16A; FCGR3; IGFR3; FCR-10; FCRIII; FCGRIII; FCRIIIA; FCGR3A | 1 | −158V/F | 12834496 | Loos BG, Leppers-Van de Straat FG, Van de Winkel JG, Van der Velden U. Fcgamma receptor polymorphisms in relation to periodontitis. J Clin Periodontol. 2003 Jul;30(7):595-602. |
| FCGR3A | AgP | 30 | 77 | ns | None | Chinese | CC | N | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | CD16; FCG3; CD16A; FCGR3; IGFR3; FCR-10; FCRIII; FCGRIII; FCRIIIA; FCGR3A | 1 | −158V/F | 19221562 | An N, Ou-Yang XY, Cao CF, Ye J, Hui RT. [Association of Fc gamma receptors IIIA gene polymorphisms with the susceptibility to periodontitis in Chinese patients]. Beijing Da Xue Xue Bao. 2009 Feb 18;41(1):40-3. Chinese. |
| FCGR3A | AgP | 9 | 41 | ns | None | Japanese | CC | N | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | CD16; FCG3; CD16A; FCGR3; IGFR3; FCR-10; FCRIII; FCGRIII; FCRIIIA; FCGR3A | 1 | H/R131 | 19184295 | Shimomura-Kuroki J, Yamashita K, Shimooka S. Tannerella forsythia and the HLA-DQB1 allele are associated with susceptibility to periodontal disease in Japanese adolescents. Odontology. 2009 Jan;97(1):32-7. Epub 2009 Jan 29. |
| FCGR3A | AgP | 224 | 455 | ns | None | Mixed | CC | EGS | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | CD16; FCG3; CD16A; FCGR3; IGFR3; FCR-10; FCRIII; FCGRIII; FCRIIIA; FCGR3A | 1 | 559 G/T | 16899095 | Nibali L, Parkar M, Brett P, Knight J, Tonetti MS, Griffiths GS. NADPH oxidase (CYBA) and FcgammaR polymorphisms as risk factors for aggressive periodontitis: a case-control association study. J Clin Periodontol. 2006 Aug;33(8):529-39. |
| FCGR3A | AgP | 32 | 104 | ns | None | Japanese | CC | N | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | CD16; FCG3; CD16A; FCGR3; IGFR3; FCR-10; FCRIII; FCGRIII; FCRIIIA; FCGR3A | 1 | n/a | 14647193 | Yasuda K, Sugita N, Kobayashi T, Yamamoto K, Yoshie H. FcgammaRIIB gene polymorphisms in Japanese periodontitis patients. Genes Immun. 2003 Dec;4(8):541-6. |
| FCGR3A | AgP | 38 | 142 | ns | None | Japanese | CC | N | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | CD16; FCG3; CD16A; FCGR3; IGFR3; FCR-10; FCRIII; FCGRIII; FCRIIIA; FCGR3A | 1 | −158V/F | 11022771 | Kobayashi T, Sugita N, van der Pol WL, Nunokawa Y, Westerdaal NA, Yamamoto K, van de Winkel JG, Yoshie H. The Fcgamma receptor genotype as a risk factor for generalized early-onset periodontitis in Japanese patients. J Periodontol. 2000 Sep;71(9):1425-32. |
| FCGR3A | AgP | 48 | 115 | ns | None | African American | CC | N | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | CD16; FCG3; CD16A; FCGR3; IGFR3; FCR-10; FCRIII; FCGRIII; FCRIIIA; FCGR3A | n/a | 158V/F | 12027254 | Fu Y, Korostoff JM, Fine DH, Wilson ME. Fc gamma receptor genes as risk markers for localized aggressive periodontitis in African-Americans. J Periodontol. 2002 May;73(5):517-23. |
| FCGR3A | AgP+CP | 319 | 622 | ns | None | Japanese | CC | AGS | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | CD16; FCG3; CD16A; FCGR3; IGFR3; FCR-10; FCRIII; FCGRIII; FCRIIIA; FCGR3A | 1 | 559 | 19892918 | Kobayashi T, Nagata T, Murakami S, Takashiba S, Kurihara H, Izumi Y, Numabe Y, Watanabe H, Kataoka M, Nagai A, Hayashi J, Ohyama H, Okamatsu Y, Inagaki Y, Tai H, Yoshie H. Genetic risk factors for periodontitis in a Japanese population. J Dent Res. 2009 Dec;88(12):1137-41. |
| FCGR3A | CP | 100 | 204 | 0.009 | Weak | Japanese | CC | N | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | CD16; FCG3; CD16A; FCGR3; IGFR3; FCR-10; FCRIII; FCGRIII; FCRIIIA; FCGR3A | 1 | −158V/F | 10444269 | Sugita N, Yamamoto K, Kobayashi T, Van Der Pol W, Horigome T, Yoshie H, Van De Winkel JG, Hara K. Relevance of Fc gamma RIIIa-158V-F polymorphism to recurrence of adult periodontitis in Japanese patients. Clin Exp Immunol. 1999 Aug;117(2):350-4. |
| FCGR3A | CP | 49 | 98 | 0.01 | Weak | German | CC | AS | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | CD16; FCG3; CD16A; FCGR3; IGFR3; FCR-10; FCRIII; FCGRIII; FCRIIIA; FCGR3A | 1 | −158V/F | 11528518 | Meisel P, Carlsson LE, Sawaf H, Fanghaenel J, Greinacher A, Kocher T. Polymorphisms of Fc gamma-receptors RIIa, RIIIa, and RIIIb in patients with adult periodontal diseases. Genes Immun. 2001 Aug;2(5):258-62. |
| FCGR3A | CP | 89 | 153 | 0.028 | Weak | Japanese | CC | S | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | CD16; FCG3; CD16A; FCGR3; IGFR3; FCR-10; FCRIII; FCGRIII; FCRIIIA; FCGR3A | 1 | −158V/F | 11699473 | Kobayashi T, Yamamoto K, Sugita N, van der Pol WL, Yasuda K, Kaneko S, van de Winkel JG, Yoshie H. The Fc gamma receptor genotype as a severity factor for chronic periodontitis in Japanese patients. J Periodontol. 2001 Oct;72(10):1324-31. |
| FCGR3A | CP | 131 | 178 | ns | None | Chinese | CC | N | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | CD16; FCG3; CD16A; FCGR3; IGFR3; FCR-10; FCRIII; FCGRIII; FCRIIIA; FCGR3A | 1 | −158V/F | 19221562 | An N, Ou-Yang XY, Cao CF, Ye J, Hui RT. [Association of Fc gamma receptors IIIA gene polymorphisms with the susceptibility to periodontitis in Chinese patients]. Beijing Da Xue Xue Bao. 2009 Feb 18;41(1):40-3. Chinese. |
| FCGR3A | CP | 100 | 200 | ns | None | Japanese | CC | N | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | CD16; FCG3; CD16A; FCGR3; IGFR3; FCR-10; FCRIII; FCGRIII; FCRIIIA; FCGR3A | 1 | 559 G/T | 18052703 | Kobayashi T, Ito S, Kuroda T, Yamamoto K, Sugita N, Narita I, Sumida T, Gejyo F, Yoshie H. The interleukin-1 and Fcgamma receptor gene polymorphisms in Japanese patients with rheumatoid arthritis and periodontitis. J Periodontol. 2007 Dec;78(12):2311-8. |
| FCGR3A | CP | 72 | 144 | ns | None | Japanese | CC | N | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | CD16; FCG3; CD16A; FCGR3; IGFR3; FCR-10; FCRIII; FCGRIII; FCRIIIA; FCGR3A | 1 | n/a | 14647193 | Yasuda K, Sugita N, Kobayashi T, Yamamoto K, Yoshie H. FcgammaRIIB gene polymorphisms in Japanese periodontitis patients. Genes Immun. 2003 Dec;4(8):541-6. |
| FCGR3A | CP | 56 | 117 | ns | None | Caucasian | CC | S | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | CD16; FCG3; CD16A; FCGR3; IGFR3; FCR-10; FCRIII; FCGRIII; FCRIIIA; FCGR3A | 1 | −158V/F | 12834496 | Loos BG, Leppers-Van de Straat FG, Van de Winkel JG, Van der Velden U. Fcgamma receptor polymorphisms in relation to periodontitis. J Clin Periodontol. 2003 Jul;30(7):595-602. |
| FCGR3A | CP | 117 | 558 | ns | None | German | CC | S | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | CD16; FCG3; CD16A; FCGR3; IGFR3; FCR-10; FCRIII; FCGRIII; FCRIIIA; FCGR3A | 1 | −158V/F | 19570260 | Meisel P, Heins G, Carlsson L, Giebel J, John U, Schwahn C, Kocher T. Impact of Genetic Polymorphisms on the Smoking-related Risk of Periodontal Disease: the Population-based Study SHIP. Tob Induc Dis. 2003 Sep 15;1(3):197. |
| FCGR3A | CP | 83 | 187 | ns | None | Japanese | CC | N | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | CD16; FCG3; CD16A; FCGR3; IGFR3; FCR-10; FCRIII; FCGRIII; FCRIIIA; FCGR3A | 1 | −158V/F | 11022771 | Kobayashi T, Sugita N, van der Pol WL, Nunokawa Y, Westerdaal NA, Yamamoto K, van de Winkel JG, Yoshie H. The Fcgamma receptor genotype as a risk factor for generalized early-onset periodontitis in Japanese patients. J Periodontol. 2000 Sep;71(9):1425-32. |
| FCGR3A | CP&RA | 86 | 186 | ns | None | Japanese | CC | N | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | CD16; FCG3; CD16A; FCGR3; IGFR3; FCR-10; FCRIII; FCGRIII; FCRIIIA; FCGR3A | 1 | 559 G/T | 18052703 | Kobayashi T, Ito S, Kuroda T, Yamamoto K, Sugita N, Narita I, Sumida T, Gejyo F, Yoshie H. The interleukin-1 and Fcgamma receptor gene polymorphisms in Japanese patients with rheumatoid arthritis and periodontitis. J Periodontol. 2007 Dec;78(12):2311-8. |
| FCGR3A | CP&SLE | 42 | 84 | ns | None | Japanese | CC | N | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | CD16; FCG3; CD16A; FCGR3; IGFR3; FCR-10; FCRIII; FCGRIII; FCRIIIA; FCGR3A | 1 | −158V/F | 12710759 | Kobayashi T, Ito S, Yamamoto K, Hasegawa H, Sugita N, Kuroda T, Kaneko S, Narita I, Yasuda K, Nakano M, Gejyo F, Yoshie H. Risk of periodontitis in systemic lupus erythematosus is associated with Fcgamma receptor polymorphisms. J Periodontol. 2003 Mar;74(3):378-84. |
| FCGR3A | CP&SLE | 42 | 60 | ns | None | Japanese | CC | N | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | CD16; FCG3; CD16A; FCGR3; IGFR3; FCR-10; FCRIII; FCGRIII; FCRIIIA; FCGR3A | 1 | −158V/F | 12710759 | Kobayashi T, Ito S, Yamamoto K, Hasegawa H, Sugita N, Kuroda T, Kaneko S, Narita I, Yasuda K, Nakano M, Gejyo F, Yoshie H. Risk of periodontitis in systemic lupus erythematosus is associated with Fcgamma receptor polymorphisms. J Periodontol. 2003 Mar;74(3):378-84. |
| FCGR3A.FCGR3B | CP | 89 | 153 | 0.0009 | Moderate | Japanese | CC | S | n/a | n/a | 2 | Multiple Genes | 11699473 | Kobayashi T, Yamamoto K, Sugita N, van der Pol WL, Yasuda K, Kaneko S, van de Winkel JG, Yoshie H. The Fc gamma receptor genotype as a severity factor for chronic periodontitis in Japanese patients. J Periodontol. 2001 Oct;72(10):1324-31. |
| FCGR3A.FCGR3B | CP | 100 | 200 | ns | None | Japanese | CC | N | n/a | n/a | 2 | Multiple Genes | 18052703 | Kobayashi T, Ito S, Kuroda T, Yamamoto K, Sugita N, Narita I, Sumida T, Gejyo F, Yoshie H. The interleukin-1 and Fcgamma receptor gene polymorphisms in Japanese patients with rheumatoid arthritis and periodontitis. J Periodontol. 2007 Dec;78(12):2311-8. |
| FCGR3A.FCGR3B | CP&RA | 86 | 186 | ns | None | Japanese | CC | N | n/a | n/a | 2 | Multiple Genes | 18052703 | Kobayashi T, Ito S, Kuroda T, Yamamoto K, Sugita N, Narita I, Sumida T, Gejyo F, Yoshie H. The interleukin-1 and Fcgamma receptor gene polymorphisms in Japanese patients with rheumatoid arthritis and periodontitis. J Periodontol. 2007 Dec;78(12):2311-8. |
| FCGR3B | AgP | 42 | 97 | 0.006 | Weak | Japanese | CC | N | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 1 | NA1/NA2 | 11808760 | Yoshihara A, Sugita N, Yamamoto K, Kobayashi T, Miyazaki H, Yoshi H. Analysis of vitamin D and Fcgamma receptor polymorphisms in Japanese patients with generalized early-onset periodontitis. J Dent Res. 2001 Dec;80(12):2051-4. |
| FCGR3B | AgP | 32 | 104 | 0.016 | Weak | Japanese | CC | N | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 1 | NA1/NA2 | 14647193 | Yasuda K, Sugita N, Kobayashi T, Yamamoto K, Yoshie H. FcgammaRIIB gene polymorphisms in Japanese periodontitis patients. Genes Immun. 2003 Dec;4(8):541-6. |
| FCGR3B | AgP | 224 | 455 | 0.019 | Weak | Mixed | CC | EGS | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 2 | NA1/NA2 | 16899095 | Nibali L, Parkar M, Brett P, Knight J, Tonetti MS, Griffiths GS. NADPH oxidase (CYBA) and FcgammaR polymorphisms as risk factors for aggressive periodontitis: a case-control association study. J Clin Periodontol. 2006 Aug;33(8):529-39. |
| FCGR3B | AgP | 38 | 142 | 0.02 | Weak | Japanese | CC | N | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 1 | NA1/NA2 | 11022771 | Kobayashi T, Sugita N, van der Pol WL, Nunokawa Y, Westerdaal NA, Yamamoto K, van de Winkel JG, Yoshie H. The Fcgamma receptor genotype as a risk factor for generalized early-onset periodontitis in Japanese patients. J Periodontol. 2000 Sep;71(9):1425-32. |
| FCGR3B | AgP | 48 | 115 | 0.024 | Weak | African American | CC | N | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | n/a | NA1/NA2 | 12027254 | Fu Y, Korostoff JM, Fine DH, Wilson ME. Fc gamma receptor genes as risk markers for localized aggressive periodontitis in African-Americans. J Periodontol. 2002 May;73(5):517-23. |
| FCGR3B | AgP | 31 | 80 | <0.001 | Weak | Brazilian | CC | EG | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 1 | NA1/NA2 | 16805673 | de Souza RC, Colombo AP. Distribution of FcgammaRIIa and FcgammaRIIIb genotypes in patients with generalized aggressive periodontitis. J Periodontol. 2006 Jul;77(7):1120-8. |
| FCGR3B | AgP | 21 | 47 | <0.05 | Weak | Chinese | CC | N | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 1 | NA1/NA2 | 12839649 | Zhang HX, Xie H, Ren TG. [Relevance of FcgammaRIIIb genotype, IgG G2m(23) factor to the susceptibility of aggressive periodontitis]. Zhonghua Kou Qiang Yi Xue Za Zhi. 2003 Mar;38(2):129-31. Chinese. |
| FCGR3B | AgP | 30 | 104 | ns | None | Taiwanese | CC | N | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 1 | NA1/NA2 | 14761117 | Chung HY, Lu HC, Chen WL, Lu CT, Yang YH, Tsai CC. Gm (23) allotypes and Fcgamma receptor genotypes as risk factors for various forms of periodontitis. J Clin Periodontol. 2003 Nov;30(11):954-60. |
| FCGR3B | AgP | 12 | 73 | ns | None | Caucasian | CC | S | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 1 | NA1/NA2 | 12834496 | Loos BG, Leppers-Van de Straat FG, Van de Winkel JG, Van der Velden U. Fcgamma receptor polymorphisms in relation to periodontitis. J Clin Periodontol. 2003 Jul;30(7):595-602. |
| FCGR3B | AgP | 33 | 60 | sig | Weak | Chinese | CC | N | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 1 | NA1/NA2 | 11776881 | Fu Y, Cao C, Wang S. [Relevance of Fc gamma R polymorphism to the susceptibility of early-onset periodontitis]. Zhonghua Kou Qiang Yi Xue Za Zhi. 1999 Nov;34(6):364-6. Chinese. |
| FCGR3B | AgP+CP | 319 | 622 | ns | None | Japanese | CC | AGS | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 1 | NA1/NA2 | 19892918 | Kobayashi T, Nagata T, Murakami S, Takashiba S, Kurihara H, Izumi Y, Numabe Y, Watanabe H, Kataoka M, Nagai A, Hayashi J, Ohyama H, Okamatsu Y, Inagaki Y, Tai H, Yoshie H. Genetic risk factors for periodontitis in a Japanese population. J Dent Res. 2009 Dec;88(12):1137-41. |
| FCGR3B | CP | 100 | 205 | 0.0003 | Moderate | Japanese | CC | N | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 1 | NA1/NA2 | 9284119 | Kobayashi T, Westerdaal NA, Miyazaki A, van der Pol WL, Suzuki T, Yoshie H, van de Winkel JG, Hara K. Relevance of immunoglobulin G Fc receptor polymorphism to recurrence of adult periodontitis in Japanese patients. Infect Immun. 1997 Sep;65(9):3556-60. |
| FCGR3B | CP | 83 | 187 | 0.009 | Weak | Japanese | CC | N | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 1 | NA1/NA2 | 11022771 | Kobayashi T, Sugita N, van der Pol WL, Nunokawa Y, Westerdaal NA, Yamamoto K, van de Winkel JG, Yoshie H. The Fcgamma receptor genotype as a risk factor for generalized early-onset periodontitis in Japanese patients. J Periodontol. 2000 Sep;71(9):1425-32. |
| FCGR3B | CP | 73 | 119 | 0.03 | Weak | Japanese | CC | N | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 1 | NA1/NA2 | 11379895 | Sugita N, Kobayashi T, Ando Y, Yoshihara A, Yamamoto K, van de Winkel JG, Miyazaki H, Yoshie H. Increased frequency of FcgammaRIIIb-NA1 allele in periodontitis-resistant subjects in an elderly Japanese population. J Dent Res. 2001 Mar;80(3):914-8. |
| FCGR3B | CP | n/a | 164 | <0.001 | Moderate | Japanese | Longitudinal | AS | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 1 | NA1/NA2 | 15974849 | Yoshihara A, Sugita N, Yamamoto K, Kobayashi T, Hirotomi T, Ogawa H, Miyazaki H, Yoshie H. FcgammaRIIIb genotypes and smoking in periodontal disease progression among community-dwelling older adults in Japan. J Periodontol. 2005 Feb;76(2):250-5. |
| FCGR3B | CP | 113 | 221 | ns | None | Japanese | CC | N | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 1 | NA1/NA2 | 18321309 | Komatsu Y, Galicia JC, Kobayashi T, Yamazaki K, Yoshie H. Association of interleukin-1 receptor antagonist +2018 gene polymorphism with Japanese chronic periodontitis patients using a novel genotyping method. Int J Immunogenet. 2008 Apr;35(2):165-70. |
| FCGR3B | CP | 100 | 200 | ns | None | Japanese | CC | N | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 1 | NA1/NA2 | 18052703 | Kobayashi T, Ito S, Kuroda T, Yamamoto K, Sugita N, Narita I, Sumida T, Gejyo F, Yoshie H. The interleukin-1 and Fcgamma receptor gene polymorphisms in Japanese patients with rheumatoid arthritis and periodontitis. J Periodontol. 2007 Dec;78(12):2311-8. |
| FCGR3B | CP | 132 | 205 | ns | None | Caucasian | CC | N | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 1 | NA1/NA2 | 16889631 | Wolf DL, Neiderud AM, Hinckley K, Dahlén G, van de Winkel JG, Papapanou PN. Fcgamma receptor polymorphisms and periodontal status: a prospective follow-up study. J Clin Periodontol. 2006 Oct;33(10):691-8. Epub 2006 Aug 3. |
| FCGR3B | CP | 72 | 144 | ns | None | Japanese | CC | N | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 1 | NA1/NA2 | 14647193 | Yasuda K, Sugita N, Kobayashi T, Yamamoto K, Yoshie H. FcgammaRIIB gene polymorphisms in Japanese periodontitis patients. Genes Immun. 2003 Dec;4(8):541-6. |
| FCGR3B | CP | 56 | 117 | ns | None | Caucasian | CC | S | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 1 | NA1/NA2 | 12834496 | Loos BG, Leppers-Van de Straat FG, Van de Winkel JG, Van der Velden U. Fcgamma receptor polymorphisms in relation to periodontitis. J Clin Periodontol. 2003 Jul;30(7):595-602. |
| FCGR3B | CP | 117 | 558 | ns | None | German | CC | S | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 1 | NA1/NA2 | 19570260 | Meisel P, Heins G, Carlsson L, Giebel J, John U, Schwahn C, Kocher T. Impact of Genetic Polymorphisms on the Smoking-related Risk of Periodontal Disease: the Population-based Study SHIP. Tob Induc Dis. 2003 Sep 15;1(3):197. |
| FCGR3B | CP | 52 | 107 | ns | None | Japanese | CC | N | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 1 | NA1/NA2 | 11808760 | Yoshihara A, Sugita N, Yamamoto K, Kobayashi T, Miyazaki H, Yoshi H. Analysis of vitamin D and Fcgamma receptor polymorphisms in Japanese patients with generalized early-onset periodontitis. J Dent Res. 2001 Dec;80(12):2051-4. |
| FCGR3B | CP | 89 | 153 | ns | None | Japanese | CC | S | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 1 | NA1/NA2 | 11699473 | Kobayashi T, Yamamoto K, Sugita N, van der Pol WL, Yasuda K, Kaneko S, van de Winkel JG, Yoshie H. The Fc gamma receptor genotype as a severity factor for chronic periodontitis in Japanese patients. J Periodontol. 2001 Oct;72(10):1324-31. |
| FCGR3B | CP | 49 | 98 | ns | None | German | CC | AS | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 1 | NA1/NA2 | 11528518 | Meisel P, Carlsson LE, Sawaf H, Fanghaenel J, Greinacher A, Kocher T. Polymorphisms of Fc gamma-receptors RIIa, RIIIa, and RIIIb in patients with adult periodontal diseases. Genes Immun. 2001 Aug;2(5):258-62. |
| FCGR3B | CP | 86 | 113 | ns | None | n/a | CC | S | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 1 | NA1/NA2 | 9667480 | Colombo AP, Eftimiadi C, Haffajee AD, Cugini MA, Socransky SS. Serum IgG2 level, Gm(23) allotype and FcgammaRIIa and FcgammaRIIIb receptors in refractory periodontal disease. J Clin Periodontol. 1998 Jun;25(6):465-74. |
| FCGR3B | CP&RA | 86 | 186 | ns | None | Japanese | CC | N | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 1 | NA1/NA2 | 18052703 | Kobayashi T, Ito S, Kuroda T, Yamamoto K, Sugita N, Narita I, Sumida T, Gejyo F, Yoshie H. The interleukin-1 and Fcgamma receptor gene polymorphisms in Japanese patients with rheumatoid arthritis and periodontitis. J Periodontol. 2007 Dec;78(12):2311-8. |
| FCGR3B | CP&SLE | 42 | 84 | ns | None | Japanese | CC | N | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 1 | NA1/NA2 | 12710759 | Kobayashi T, Ito S, Yamamoto K, Hasegawa H, Sugita N, Kuroda T, Kaneko S, Narita I, Yasuda K, Nakano M, Gejyo F, Yoshie H. Risk of periodontitis in systemic lupus erythematosus is associated with Fcgamma receptor polymorphisms. J Periodontol. 2003 Mar;74(3):378-84. |
| FCGR3B | CP&SLE | 42 | 60 | ns | None | Japanese | CC | N | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | CD16; FCG3; CD16b; FCGR3; FCGR3B | 1 | NA1/NA2 | 12710759 | Kobayashi T, Ito S, Yamamoto K, Hasegawa H, Sugita N, Kuroda T, Kaneko S, Narita I, Yasuda K, Nakano M, Gejyo F, Yoshie H. Risk of periodontitis in systemic lupus erythematosus is associated with Fcgamma receptor polymorphisms. J Periodontol. 2003 Mar;74(3):378-84. |
| FCN1 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Ficolin (collagen/fibrinogen domain containing) 1 | FCNM; FCN1 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| FGA | CP | 22 | 41 | ns | None | Japanese | CC | N | Fibrinogen alpha chain | Fib2; MGC119422; MGC119423; MGC119425; FGA | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| FGB | AgP+CP | 319 | 622 | ns | None | Japanese | CC | AGS | Fibrinogen beta chain | MGC104327; MGC120405; FGB | 1 | −455 | 19892918 | Kobayashi T, Nagata T, Murakami S, Takashiba S, Kurihara H, Izumi Y, Numabe Y, Watanabe H, Kataoka M, Nagai A, Hayashi J, Ohyama H, Okamatsu Y, Inagaki Y, Tai H, Yoshie H. Genetic risk factors for periodontitis in a Japanese population. J Dent Res. 2009 Dec;88(12):1137-41. |
| FGB | CP | n/a | 121 | 0.008 | Weak | Chinese | CC | N | Fibrinogen beta chain | MGC104327; MGC120405; FGB | 1 | −455 G/A | 18683729 | Ge S, Wu YF, Liu TJ, He QM, Zhao L, Meng S. [Correlation between levels of fibrinogen, beta455 g/A fibrinogen gene polymorphism and chronic periodontitis]. Zhonghua Kou Qiang Yi Xue Za Zhi. 2008 Feb;43(2):87-91. Chinese. |
| FGB | CP | 79 | 154 | 0.01 | Weak | Mixed | CC | N | Fibrinogen beta chain | MGC104327; MGC120405; FGB | 1 | HaeIII, -455 | 12710752 | Sahingur SE, Sharma A, Genco RJ, De Nardin E. Association of increased levels of fibrinogen and the -455G/A fibrinogen gene polymorphism with chronic periodontitis. J Periodontol. 2003 Mar;74(3):329-37. |
| FGF1 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Fibroblast growth factor 1 (acidic) | AFGF; ECGF; FGFA; ECGFA; ECGFB; HBGF1; GLIO703; ECGF-beta; FGF-alpha; FGF1 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| FGF10 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Fibroblast growth factor 10 | FGF10 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| FGF12 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Fibroblast growth factor 12 | FHF1; FGF12B; FGF12 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| FGF12 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Fibroblast growth factor 12 | FHF1; FGF12B; FGF12 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| FGF13 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Fibroblast growth factor 13 | FGF2; FHF2; FHF-2; FGF13 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| FGF2 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Fibroblast growth factor 2 (basic) | BFGF; FGFB; HBGF-2; FGF2 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| FGF21 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Fibroblast growth factor 21 | FGF21 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| FGF6 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Fibroblast growth factor 6 | HST2; HBGF-6; FGF6 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| FGF9 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Fibroblast growth factor 9 (glia-activating factor) | GAF; SYNS3; HBFG-9; MGC119914; MGC119915; FGF9 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| FGFRL1 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Fibroblast growth factor receptor-like 1 | FHFR; FGFR5; FGFRL1 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| FGL1 | CP | 22 | 41 | ns | None | Japanese | CC | N | Fibrinogen-like 1 | HFREP1; HP-041; LFIRE1; MGC12455; FGL1 | 2 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| FGL2 | CP | 22 | 41 | 0.007 | Weak | Japanese | CC | N | Fibrinogen-like 2 | T49; pT49; FGL2 | 1 | IMS-JST003521 | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| FN1 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Fibronectin 1 | FN; CIG; FNZ; MSF; ED-B; FINC; GFND; LETS; GFND2; DKFZp686H0342; DKFZp686I1370; DKFZp686F10164; DKFZp686O13149; FN1 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| FOXK2 | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Forkhead box K2 | ILF; ILF1; ILF-1; FOXK2 | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| FPR1 | AgP | 49 | 422 | 0.00002 | Moderate | Japanese | CC | N | Formyl peptide receptor 1 | FPR; FMLP; FPR1 | 3 | −12915C>T.301G>C.546C>A | 17927965 | Gunji T, Onouchi Y, Nagasawa T, Katagiri S, Watanabe H, Kobayashi H, Arakawa S, Noguchi K, Hata A, Izumi Y, Ishikawa I. Functional polymorphisms of the FPR1 gene and aggressive periodontitis in Japanese. Biochem Biophys Res Commun. 2007 Dec 7;364(1):7-13. Epub 2007 Oct 2. |
| FPR1 | AgP | 30 | 63 | 0.0005 | Weak | African American | CC | N | Formyl peptide receptor 1 | FPR; FMLP; FPR1 | 6 | 348T>C | 19254133 | Maney P, Emecen P, Mills JS, Walters JD. Neutrophil formylpeptide receptor single nucleotide polymorphism 348T>C in aggressive periodontitis. J Periodontol. 2009 Mar;80(3):492-8. |
| FPR1 | AgP | 30 | 50 | 0.0005 | Weak | Mixed | CC | E | Formyl peptide receptor 1 | FPR; FMLP; FPR1 | 3 | Cys126Try or Phe110Ser | 10534074 | Gwinn MR, Sharma A, De Nardin E. Single nucleotide polymorphisms of the N-formyl peptide receptor in localized juvenile periodontitis. J Periodontol. 1999 Oct;70(10):1194-201. |
| FPR1 | AgP | 49 | 422 | 0.00098 | Moderate | Japanese | CC | N | Formyl peptide receptor 1 | FPR; FMLP; FPR1 | 30 | −12915C>T | 17927965 | Gunji T, Onouchi Y, Nagasawa T, Katagiri S, Watanabe H, Kobayashi H, Arakawa S, Noguchi K, Hata A, Izumi Y, Ishikawa I. Functional polymorphisms of the FPR1 gene and aggressive periodontitis in Japanese. Biochem Biophys Res Commun. 2007 Dec 7;364(1):7-13. Epub 2007 Oct 2. |
| FPR1 | AgP | 111 | 226 | 0.0018 | Weak | Mixed | CC | E | Formyl peptide receptor 1 | FPR; FMLP; FPR1 | 6 | N192K | 12595898 | Zhang Y, Syed R, Uygar C, Pallos D, Gorry MC, Firatli E, Cortelli JR, VanDyke TE, Hart PS, Feingold E, Hart TC. Evaluation of human leukocyte N-formylpeptide receptor (FPR1) SNPs in aggressive periodontitis patients. Genes Immun. 2003 Jan;4(1):22-9. |
| FPR1 | AgP | 30 | 63 | 0.0036 | Weak | African American | CC | N | Formyl peptide receptor 1 | FPR; FMLP; FPR1 | 4 | Haplotype | 19254133 | Maney P, Emecen P, Mills JS, Walters JD. Neutrophil formylpeptide receptor single nucleotide polymorphism 348T>C in aggressive periodontitis. J Periodontol. 2009 Mar;80(3):492-8. |
| FPR1 | AgP | 37 | 75 | 0.017 | Weak | African American | CC | N | Formyl peptide receptor 1 | FPR; FMLP; FPR1 | 12 | 348T>C | 19722801 | Maney P, Walters JD. Formylpeptide Receptor Single Nucleotide Polymorphism 348T>C and Its Relationship to Polymorphonuclear Leukocyte Chemotaxis in Aggressive Periodontitis. J Periodontol. 2009 Sep;80(9):1498-505. |
| FPR1 | AgP | 37 | 75 | ns | None | African American | CC | N | Formyl peptide receptor 1 | FPR; FMLP; FPR1 | 4 | Haplotype | 19722801 | Maney P, Walters JD. Formylpeptide Receptor Single Nucleotide Polymorphism 348T>C and Its Relationship to Polymorphonuclear Leukocyte Chemotaxis in Aggressive Periodontitis. J Periodontol. 2009 Sep;80(9):1498-505. |
| FPR1 | AgP | 75 | 138 | ns | None | Turkish | CC | N | Formyl peptide receptor 1 | FPR; FMLP; FPR1 | 6 | Multiple SNPs | 19254133 | Maney P, Emecen P, Mills JS, Walters JD. Neutrophil formylpeptide receptor single nucleotide polymorphism 348T>C in aggressive periodontitis. J Periodontol. 2009 Mar;80(3):492-8. |
| FPR1 | AgP | 75 | 138 | ns | None | Turkish | CC | N | Formyl peptide receptor 1 | FPR; FMLP; FPR1 | 4 | Haplotype | 19254133 | Maney P, Emecen P, Mills JS, Walters JD. Neutrophil formylpeptide receptor single nucleotide polymorphism 348T>C in aggressive periodontitis. J Periodontol. 2009 Mar;80(3):492-8. |
| FPR1 | AgP | 224 | 455 | ns | None | Mixed | CC | EGS | Formyl peptide receptor 1 | FPR; FMLP; FPR1 | 3 | Multiple SNPs | 16899095 | Nibali L, Parkar M, Brett P, Knight J, Tonetti MS, Griffiths GS. NADPH oxidase (CYBA) and FcgammaR polymorphisms as risk factors for aggressive periodontitis: a case-control association study. J Clin Periodontol. 2006 Aug;33(8):529-39. |
| FPR1 | AgP+CP | 319 | 622 | ns | None | Japanese | CC | AGS | Formyl peptide receptor 1 | FPR; FMLP; FPR1 | 2 | n/a | 19892918 | Kobayashi T, Nagata T, Murakami S, Takashiba S, Kurihara H, Izumi Y, Numabe Y, Watanabe H, Kataoka M, Nagai A, Hayashi J, Ohyama H, Okamatsu Y, Inagaki Y, Tai H, Yoshie H. Genetic risk factors for periodontitis in a Japanese population. J Dent Res. 2009 Dec;88(12):1137-41. |
| FPR1 | CP | 22 | 41 | ns | None | Japanese | CC | N | Formyl peptide receptor 1 | FPR; FMLP; FPR1 | 2 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| GAL3ST1 | CP | 22 | 41 | ns | None | Japanese | CC | N | Galactose-3-O-sulfotransferase 1 | CST; GAL3ST1 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| GBP2 | CP | 22 | 41 | ns | None | Japanese | CC | N | Guanylate binding protein 2, interferon-inducible | GBP2 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| GC | AgP+CP | 146 | 228 | ns | None | Mixed | Fam | N | Group-specific component (vitamin D binding protein) | DBP; VDBG; VDBP; DBP/GC; GC | 1 | n/a | 8100208 | Hart TC, Marazita ML, McCanna KM, Schenkein HA, Diehl SR. Reevaluation of the chromosome 4q candidate region for early onset periodontitis. Hum Genet. 1993 Jun;91(5):416-22. |
| GCK | CP | 22 | 41 | ns | None | Japanese | CC | N | Glucokinase (hexokinase 4) | GK; GLK; HK4; HHF3; HKIV; HXKP; MODY2; GCK | 3 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| GHRHR | CP | 22 | 41 | ns | None | Japanese | CC | N | Growth hormone releasing hormone receptor | GRFR; GHRFR; IGHD1B; GHRHRpsv; GHRHR | 2 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| GIP | CP | 22 | 41 | ns | None | Japanese | CC | N | Gastric inhibitory polypeptide | GIP | 2 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| GIPR | CP | 22 | 41 | ns | None | Japanese | CC | N | Gastric inhibitory polypeptide receptor | MGC126722; GIPR | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| GLP1R | CP | 22 | 41 | ns | None | Japanese | CC | N | GlucaGon-like peptide 1 receptor | MGC138331; GLP1R | 2 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| GLP2R | CP | 22 | 41 | ns | None | Japanese | CC | N | Glucagon-like peptide 2 receptor | GLP2R | 3 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| GLT6D1 | AgP | 447 | 1787 | 5.51E-09 | Strong | Caucasian | CC | ADGS | Glycosyltransferase 6 domain containing 1 | GT6M7; GLTDC1; GLT6D1 | GWAS | rs1537415 | 19897590 | Schaefer AS, Richter GM, Nothnagel M, Manke T, Dommisch H, Jacobs G, Arlt A, Rosenstiel P, Noack B, Groessner-Schreiber B, Jepsen S, Loos BG, Schreiber S. A genome-wide association study identifies GLT6D1 as a susceptibility locus for periodontitis. Hum Mol Genet. 2009 Dec 1. |
| GNB3 | CP | 22 | 41 | ns | None | Japanese | CC | N | Guanine nucleotide binding protein (G protein), beta polypeptide 3 | GNB3 | 2 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| GNRH1 | CP | 22 | 41 | 0.0008 | Weak | Japanese | CC | N | Gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) | GRH; GNRH; LHRH; LNRH; GNRH1 | 1 | IMS-JST057216 | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| GPR39 | CP | 22 | 41 | ns | None | Japanese | CC | N | G protein-coupled receptor 39 | MGC149541; GPR39 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| GPX3 | CP | 22 | 41 | ns | None | Japanese | CC | N | Glutathione peroxidase 3 (plasma) | GPx-P; GSHPx-3; GSHPx-P; GPX3 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| GPX5 | CP | 22 | 41 | ns | None | Japanese | CC | N | Glutathione peroxidase 5 (epididymal androgen-related protein) | GPX5 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| GRP | CP | 22 | 41 | ns | None | Japanese | CC | N | Gastrin-releasinG peptide | BN; GRP-10; proGRP; preproGRP; GRP | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| GSTM1 | AgP | 14 | 75 | 0.002 | Weak | Caucasian | CC | N | Glutathione S-transferase mu 1 | MU; H-B; GST1; GTH4; GTM1; MU-1; GSTM1-1; MGC26563; GSTM1a-1a; GSTM1b-1b; GSTM1 | 1 | n/a | 17524385 | Concolino P, Cecchetti F, D’Autilia C, Santonocito C, Di Stasio E, Zuppi C, Arcuri C, Deli G, Giardina B, Capoluongo E, Ameglio F. Association of periodontitis with GSTM1/GSTT1-null variants-a pilot study. Clin Biochem. 2007 Sep;40(13-14):939-45. Epub 2007 Apr 27. |
| GSTM1 | AgP+CP | 83 | 144 | 0.0007 | Moderate | Caucasian | CC | N | Glutathione S-transferase mu 1 | MU; H-B; GST1; GTH4; GTM1; MU-1; GSTM1-1; MGC26563; GSTM1a-1a; GSTM1b-1b; GSTM1 | 1 | n/a | 17524385 | Concolino P, Cecchetti F, D’Autilia C, Santonocito C, Di Stasio E, Zuppi C, Arcuri C, Deli G, Giardina B, Capoluongo E, Ameglio F. Association of periodontitis with GSTM1/GSTT1-null variants-a pilot study. Clin Biochem. 2007 Sep;40(13-14):939-45. Epub 2007 Apr 27. |
| GSTM1 | CP | 69 | 130 | 0.001 | Weak | Caucasian | CC | AGHS | Glutathione S-transferase mu 1 | MU; H-B; GST1; GTH4; GTM1; MU-1; GSTM1-1; MGC26563; GSTM1a-1a; GSTM1b-1b; GSTM1 | 1 | n/a | 17524385 | Concolino P, Cecchetti F, D’Autilia C, Santonocito C, Di Stasio E, Zuppi C, Arcuri C, Deli G, Giardina B, Capoluongo E, Ameglio F. Association of periodontitis with GSTM1/GSTT1-null variants-a pilot study. Clin Biochem. 2007 Sep;40(13-14):939-45. Epub 2007 Apr 27. |
| GSTM1 | CP | 115 | 241 | 0.05 | Weak | Korean | CC | S | Glutathione S-transferase mu 1 | MU; H-B; GST1; GTH4; GTM1; MU-1; GSTM1-1; MGC26563; GSTM1a-1a; GSTM1b-1b; GSTM1 | 1 | n/a | 15491310 | Kim JS, Park JY, Chung WY, Choi MA, Cho KS, Park KK. Polymorphisms in genes coding for enzymes metabolizing smoking-derived substances and the risk of periodontitis. J Clin Periodontol. 2004 Nov;31(11):959-64. |
| GSTM1.GSTT1 | AgP | 14 | 75 | <0.0001 | Moderate | Caucasian | CC | N | n/a | n/a | 2 | Multiple Genes | 17524385 | Concolino P, Cecchetti F, D’Autilia C, Santonocito C, Di Stasio E, Zuppi C, Arcuri C, Deli G, Giardina B, Capoluongo E, Ameglio F. Association of periodontitis with GSTM1/GSTT1-null variants-a pilot study. Clin Biochem. 2007 Sep;40(13-14):939-45. Epub 2007 Apr 27. |
| GSTM1.GSTT1 | AgP+CP | 83 | 144 | 0.034 | Weak | Caucasian | CC | AGHS | n/a | n/a | 2 | Multiple Genes | 17524385 | Concolino P, Cecchetti F, D’Autilia C, Santonocito C, Di Stasio E, Zuppi C, Arcuri C, Deli G, Giardina B, Capoluongo E, Ameglio F. Association of periodontitis with GSTM1/GSTT1-null variants-a pilot study. Clin Biochem. 2007 Sep;40(13-14):939-45. Epub 2007 Apr 27. |
| GSTT1 | AgP | 14 | 75 | ns | None | Caucasian | CC | N | Glutathione S-transferase theta 1 | GSTT1 | 1 | n/a | 17524385 | Concolino P, Cecchetti F, D’Autilia C, Santonocito C, Di Stasio E, Zuppi C, Arcuri C, Deli G, Giardina B, Capoluongo E, Ameglio F. Association of periodontitis with GSTM1/GSTT1-null variants-a pilot study. Clin Biochem. 2007 Sep;40(13-14):939-45. Epub 2007 Apr 27. |
| GSTT1 | AgP+CP | 83 | 144 | ns | None | Caucasian | CC | N | Glutathione S-transferase theta 1 | GSTT1 | 1 | n/a | 17524385 | Concolino P, Cecchetti F, D’Autilia C, Santonocito C, Di Stasio E, Zuppi C, Arcuri C, Deli G, Giardina B, Capoluongo E, Ameglio F. Association of periodontitis with GSTM1/GSTT1-null variants-a pilot study. Clin Biochem. 2007 Sep;40(13-14):939-45. Epub 2007 Apr 27. |
| GSTT1 | CP | 69 | 130 | ns | None | Caucasian | CC | N | Glutathione S-transferase theta 1 | GSTT1 | 1 | n/a | 17524385 | Concolino P, Cecchetti F, D’Autilia C, Santonocito C, Di Stasio E, Zuppi C, Arcuri C, Deli G, Giardina B, Capoluongo E, Ameglio F. Association of periodontitis with GSTM1/GSTT1-null variants-a pilot study. Clin Biochem. 2007 Sep;40(13-14):939-45. Epub 2007 Apr 27. |
| GSTT2 | CP | 22 | 41 | ns | None | Japanese | CC | N | Glutathione S-transferase theta 2 | MGC182032; GSTT2 | 1 | n/a | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| GZMB | CP | 22 | 41 | ns | None | Japanese | CC | N | Granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) | HLP; CCPI; CGL1; CSPB; SECT; CGL-1; CSP-B; CTLA1; CTSGL1; GZMB | 2 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| HBEGF | AgP+CP | 251 | 376 | ns | None | Japanese | CC | N | Heparin-binding EGF-like growth factor | DTR; DTS; DTSF; HEGFL; HBEGF | n/a | n/a | 15081423 | Suzuki A, Ji G, Numabe Y, Muramatsu M, Gomi K, Kanazashi M, Ogata Y, Shimizu E, Shibukawa Y, Ito A, Ito T, Sugaya A, Arai T, Yamada S, Deguchi S, Kamoi K. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004 May 7;317(3):887-92. |
| HFE | CP | 22 | 41 | ns | None | Japanese | CC | N | Hemochromatosis | HH; HFE1; HLA-H; MVCD7; MGC103790; dJ221C16.10.1; HFE | 3 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| HGF | CP | 22 | 41 | ns | None | Japanese | CC | N | Hepatocyte growth factor (hepapoietin A; scatter factor) | SF; HGFB; HPTA; F-TCF; DFNB39; HGF | 3 | Multiple SNPs | 15490304 | Suzuki A, Ji G, Numabe Y, Ishii K, Muramatsu M, Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004 Sep;92(1):43-7. |
| HLA-A | AgP | 50 | 152 | 0.007 | Weak | Caucasian | CC | N | HLAA; FLJ26655; HLA-A | n/a | A*30/31 | 12296785 | Machulla HK, Stein J, Gautsch A, Langner J, Schaller HG, Reichert S. HLA-A, B, Cw, DRB1, DRB3/4/5, DQB1 in German patients suffering from rapidly progressive periodontitis (RPP) and adult periodontitis (AP). J Clin Periodontol. 2002 Jun;29(6):573-9. | |
| HLA-A | AgP | 50 | 152 | 0.007 | Weak | Caucasian | CC | G | Major histocompatibility complex, class I, A | HLAA; FLJ26655; HLA-A | n/a | A*68/69 | 12485327 | Reichert S, Stein J, Gautsch A, Schaller HG, Machulla HK. Gender differences in HLA phenotype frequencies found in German patients with generalized aggressive periodontitis and chronic periodontitis. Oral Microbiol Immunol. 2002 Dec;17(6):360-8. |
| HLA-A | AgP | 44 | 2085 | 0.018 | Weak | Caucasian | CC | N | Major histocompatibility complex, class I, A | HLAA; FLJ26655; HLA-A | n/a | A24 | 3641478 | Klouda PT, Porter SR, Scully C, Corbin SA, Bradley BA, Smith R, Davies RM. Association between HLA-A9 and rapidly progressive periodontitis. Tissue Antigens. 1986 Sep;28(3):146-9. |
| HLA-A | AgP | 18 | 18 | 0.04 | Weak | Black | CC | N | Major histocompatibility complex, class I, A | HLAA; FLJ26655; HLA-A | 14 | A33 | 3457042 | Cogen RB, Roseman JM, Al-Joburi W, Louv WC, Acton RT, Barger BO, Go RC, Rasmussen RA. Host factors in juvenile periodontitis. J Dent Res. 1986 Mar;65(3):394-9. |
| HLA-A | AgP | 50 | 152 | 0.04 | Weak | Caucasian | CC | N | Major histocompatibility complex, class I, A | HLAA; FLJ26655; HLA-A | n/a | A*02, A*03 | 12941076 | Stein J, Reichert S, Gautsch A, Machulla HK. Are there HLA combinations typical supporting for or making resistant against aggressive and/or chronic periodontitis? J Periodontal Res. 2003 Oct;38(5):508-17. |
| HLA-A | AgP | 28 | 69 | <0.003 | Weak | Caucasian | CC | N | Major histocompatibility complex, class I, A | HLAA; FLJ26655; HLA-A | n/a | A2 | 1054359 | Kaslick RS, West TL, Chasens AI, Terasaki PI, Lazzara R, Weinberg S. Association between HL-A2 antigen and various periodontal diseases in young adults. J Dent Res. 1975 Mar-Apr;54(2):424. |
| HLA-A | AgP | 26 | 139 | <0.005 | Weak | non-Ashkenazi | CC | E | Major histocompatibility complex, class I, A | HLAA; FLJ26655; HLA-A | n/a | A9 | 15111626 | Shapira L, Eizenberg S, Sela MN, Soskolne A, Brautbar H. HLA A9 and B15 are associated with the generalized form, but not the localized form, of early-onset periodontal diseases. J Periodontol. 1994 Mar;65(3):219-23. |
| HLA-A | AgP | 49 | 89 | <0.01 | Weak | Caucasian | CC | N | Major histocompatibility complex, class I, A | HLAA; FLJ26655; HLA-A | n/a | A9 | 3163857 | Amer A, Singh G, Darke C, Dolby AE. Association between HLA antigens and periodontal disease. Tissue Antigens. 1988 Feb;31(2):53-8. |
| HLA-A | AgP | 26 | 139 | <0.05 | Weak | non-Ashkenazi | CC | E | Major histocompatibility complex, class I, A | HLAA; FLJ26655; HLA-A | n/a | B15 | 15111626 | Shapira L, Eizenberg S, Sela MN, Soskolne A, Brautbar H. HLA A9 and B15 are associated with the generalized form, but not the localized form, of early-onset periodontal diseases. J Periodontol. 1994 Mar;65(3):219-23. |
| HLA-A | AgP | 30 | 370 | <0.05 | Weak | Mixed | CC | E | Major histocompatibility complex, class I, A | HLAA; FLJ26655; HLA-A | n/a | Blank | 6155463 | Cullinan MP, Sachs J, Wolf E, Seymour GJ. The distribution of HLA-A and -B antigens in patients and their families with periodontosis. J Periodontal Res. 1980 Mar;15(2):177-84. |
| HLA-A | AgP | 19 | 60 | ns | None | Caucasian | CC | N | Major histocompatibility complex, class I, A | HLAA; FLJ26655; HLA-A | n/a | A2 | 50636 | Terasaki PI, Kaslick RS, West TL, Chasens AI. Low HL-A2 frequency and periodontitis. Tissue Antigens. 1975 May;5(4):286-8. |
| HLA-A | AgP | 19 | 60 | ns | None | Caucasian | CC | N | Major histocompatibility complex, class I, A | HLAA; FLJ26655; HLA-A | n/a | A2 | 1054359 | Kaslick RS, West TL, Chasens AI, Terasaki PI, Lazzara R, Weinberg S. Association between HL-A2 antigen and various periodontal diseases in young adults. J Dent Res. 1975 Mar-Apr;54(2):424. |
| HLA-A | AgP | 10 | 130 | ns | None | Mixed | CC | N | Major histocompatibility complex, class I, A | HLAA; FLJ26655; HLA-A | n/a | Multiple SNPs | 3498813 | Katz J, Goultschin J, Benoliel R, Brautbar C. Human leukocyte antigen (HLA) DR4. Positive association with rapidly progressing periodontitis. J Periodontol. 1987 Sep;58(9):607-10. |
| HLA-A | AgP | 60 | 3791 | sig | Weak | Turkish | CC | N | Major histocompatibility complex, class I, A | HLAA; FLJ26655; HLA-A | 13 | A9 | 8811476 | Firatli E, Kantarci A, Cebeci I, Tanyeri H, Sönmez G, Carin M, Tuncer O. Association between HLA antigens and early onset periodontitis. J Clin Periodontol. 1996 Jun;23(6):563-6. |
| HLA-A | AgP | 39 | 2006 | sig | Weak | Danish | CC | N | Major histocompatibility complex, class I, A | HLAA; FLJ26655; HLA-A | n/a | A9, A28 | 272392 | Reinholdt J, Bay I, Svejgaard A. Association between HLA-antigens and periodontal disease. J Dent Res. 1977 Oct;56(10):1261-3. |
| HLA-A | CP | 102 | 204 | 0.032 | Weak | Caucasian | CC | N | Major histocompatibility complex, class I, A | HLAA; FLJ26655; HLA-A | n/a | A*29 | 12296785 | Machulla HK, Stein J, Gautsch A, Langner J, Schaller HG, Reichert S. HLA-A, B, Cw, DRB1, DRB3/4/5, DQB1 in German patients suffering from rapidly progressive periodontitis (RPP) and adult periodontitis (AP). J Clin Periodontol. 2002 Jun;29(6):573-9. |
| HLA-A | CP | 102 | 204 | 0.032 | Weak | Caucasian | CC | N | Major histocompatibility complex, class I, A | HLAA; FLJ26655; HLA-A | n/a | A*01, A*03 | 12941076 | Stein J, Reichert S, Gautsch A, Machulla HK. Are there HLA combinations typical supporting for or making resistant against aggressive and/or chronic periodontitis? J Periodontal Res. 2003 Oct;38(5):508-17. |
| HLA-A | CP | 102 | 204 | 0.04 | Weak | Caucasian | CC | G | Major histocompatibility complex, class I, A | HLAA; FLJ26655; HLA-A | n/a | A*11 | 12485327 | Reichert S, Stein J, Gautsch A, Schaller HG, Machulla HK. Gender differences in HLA phenotype frequencies found in German patients with generalized aggressive periodontitis and chronic periodontitis. Oral Microbiol Immunol. 2002 Dec;17(6):360-8. |
| HLA-A | CP | 40 | 120 | ns | None | South Indian | CC | N | Major histocompatibility complex, class I, A | HLAA; FLJ26655; HLA-A | 1 | A9 | 17209778 | Roshna T, Thomas R, Nandakumar K, Banerjee M. A case-control study on the association of human leukocyte antigen-A*9 and -B*15 alleles with generalized aggressive periodontitis in an Indian population. J Periodontol. 2006 Dec;77(12):1954-63. |
| HLA-A | CP | 25 | 50 | ns | None | Mixed | CC | E | Major histocompatibility complex, class I, A | HLAA; FLJ26655; HLA-A | n/a | A28 | 6584591 | Goteiner D, Goldman MJ. Human lymphocyte antigen haplotype and resistance to periodontitis. J Periodontol. 1984 Mar;55(3):155-8. |
| HLA-A | CP | 28 | 69 | sig | Weak | Caucasian | CC | N | Major histocompatibility complex, class I, A | HLAA; FLJ26655; HLA-A | n/a | A2 | 50636 | Terasaki PI, Kaslick RS, West TL, Chasens AI. Low HL-A2 frequency and periodontitis. Tissue Antigens. 1975 May;5(4):286-8. |
| HLA-A | CP | 62 | 106 | sig | Weak | French | CC | N | Major histocompatibility complex, class I, A | HLAA; FLJ26655; HLA-A | n/a | A9 | 3093668 | Balndin-Texier A, Gueguin M, Fauchet R, Yardin M, Cathelineau G. [The HLA-A9 antigen and chronic periodontitis]. J Parodontol. 1986 Sep;5(3):221-7. |
| HLA-A.HLA-B | AgP | 50 | 152 | 0.034 | Weak | Caucasian | CC | N | n/a | n/a | n/a | Multiple Genes | 12941076 | Stein J, Reichert S, Gautsch A, Machulla HK. Are there HLA combinations typical supporting for or making resistant against aggressive and/or chronic periodontitis? J Periodontal Res. 2003 Oct;38(5):508-17. |
| HLA-A.HLA-B | CP | 40 | 120 | 0.011 | Weak | South Indian | CC | N | n/a | n/a | 2 | Multiple Genes | 17209778 | Roshna T, Thomas R, Nandakumar K, Banerjee M. A case-control study on the association of human leukocyte antigen-A*9 and -B*15 alleles with generalized aggressive periodontitis in an Indian population. J Periodontol. 2006 Dec;77(12):1954-63. |
| HLA-A.HLA-B | CP | 102 | 204 | 0.03 | Weak | Caucasian | CC | N | n/a | n/a | n/a | Multiple Genes | 12941076 | Stein J, Reichert S, Gautsch A, Machulla HK. Are there HLA combinations typical supporting for or making resistant against aggressive and/or chronic periodontitis? J Periodontal Res. 2003 Oct;38(5):508-17. |
| HLA-A.HLA-Cw | AgP | 50 | 152 | 0.031 | Weak | Caucasian | CC | N | n/a | n/a | n/a | Multiple Genes | 12941076 | Stein J, Reichert S, Gautsch A, Machulla HK. Are there HLA combinations typical supporting for or making resistant against aggressive and/or chronic periodontitis? J Periodontal Res. 2003 Oct;38(5):508-17. |
| HLA-B | AgP | 30 | 370 | <0.001 | Weak | Mixed | CC | E | Major histocompatibility complex, class I, B | AS; HLAB; HLAC; SPDA1; HLA-B27; HLA-B73; MGC111087; HLA-B-7301; HLA-B | n/a | BW35 | 6155463 | Cullinan MP, Sachs J, Wolf E, Seymour GJ. The distribution of HLA-A and -B antigens in patients and their families with periodontosis. J Periodontal Res. 1980 Mar;15(2):177-84. |
| HLA-B | AgP | 18 | 18 | ns | None | Black | CC | N | Major histocompatibility complex, class I, B | AS; HLAB; HLAC; SPDA1; HLA-B27; HLA-B73; MGC111087; HLA-B-7301; HLA-B | 32 | Multiple SNPs | 3457042 | Cogen RB, Roseman JM, Al-Joburi W, Louv WC, Acton RT, Barger BO, Go RC, Rasmussen RA. Host factors in juvenile periodontitis. J Dent Res. 1986 Mar;65(3):394-9. |
| HLA-B | AgP | 60 | 3791 | ns | None | Turkish | CC | N | Major histocompatibility complex, class I, B | AS; HLAB; HLAC; SPDA1; HLA-B27; HLA-B73; MGC111087; HLA-B-7301; HLA-B | 18 | Multiple SNPs | 8811476 | Firatli E, Kantarci A, Cebeci I, Tanyeri H, Sönmez G, Carin M, Tuncer O. Association between HLA antigens and early onset periodontitis. J Clin Periodontol. 1996 Jun;23(6):563-6. |
| HLA-B | AgP | 50 | 152 | ns | None | Caucasian | CC | N | Major histocompatibility complex, class I, B | AS; HLAB; HLAC; SPDA1; HLA-B27; HLA-B73; MGC111087; HLA-B-7301; HLA-B | n/a | n/a | 12296785 | Machulla HK, Stein J, Gautsch A, Langner J, Schaller HG, Reichert S. HLA-A, B, Cw, DRB1, DRB3/4/5, DQB1 in German patients suffering from rapidly progressive periodontitis (RPP) and adult periodontitis (AP). J Clin Periodontol. 2002 Jun;29(6):573-9. |
| HLA-B | AgP | 50 | 152 | ns | None | Caucasian | CC | G | Major histocompatibility complex, class I, B | AS; HLAB; HLAC; SPDA1; HLA-B27; HLA-B73; MGC111087; HLA-B-7301; HLA-B | n/a | n/a | 12485327 | Reichert S, Stein J, Gautsch A, Schaller HG, Machulla HK. Gender differences in HLA phenotype frequencies found in German patients with generalized aggressive periodontitis and chronic periodontitis. Oral Microbiol Immunol. 2002 Dec;17(6):360-8. |
| HLA-B | AgP | 50 | 152 | ns | None | Caucasian | CC | N | Major histocompatibility complex, class I, B | AS; HLAB; HLAC; SPDA1; HLA-B27; HLA-B73; MGC111087; HLA-B-7301; HLA-B | n/a | n/a | 12941076 | Stein J, Reichert S, Gautsch A, Machulla HK. Are there HLA combinations typical supporting for or making resistant against aggressive and/or chronic periodontitis? J Periodontal Res. 2003 Oct;38(5):508-17. |
| HLA-B | AgP | 49 | 89 | ns | None | Caucasian | CC | N | Major histocompatibility complex, class I, B | AS; HLAB; HLAC; SPDA1; HLA-B27; HLA-B73; MGC111087; HLA-B-7301; HLA-B | n/a | n/a | 3163857 | Amer A, Singh G, Darke C, Dolby AE. Association between HLA antigens and periodontal disease. Tissue Antigens. 1988 Feb;31(2):53-8. |
| HLA-B | AgP | 10 | 130 | ns | None | Mixed | CC | N | Major histocompatibility complex, class I, B | AS; HLAB; HLAC; SPDA1; HLA-B27; HLA-B73; MGC111087; HLA-B-7301; HLA-B | n/a | Multiple SNPs | 3498813 | Katz J, Goultschin J, Benoliel R, Brautbar C. Human leukocyte antigen (HLA) DR4. Positive association with rapidly progressing periodontitis. J Periodontol. 1987 Sep;58(9):607-10. |
| HLA-B | AgP | 44 | 2085 | ns | None | Caucasian | CC | N | Major histocompatibility complex, class I, B | AS; HLAB; HLAC; SPDA1; HLA-B27; HLA-B73; MGC111087; HLA-B-7301; HLA-B | n/a | n/a | 3641478 | Klouda PT, Porter SR, Scully C, Corbin SA, Bradley BA, Smith R, Davies RM. Association between HLA-A9 and rapidly progressive periodontitis. Tissue Antigens. 1986 Sep;28(3):146-9. |
| HLA-B | AgP | 39 | 2006 | sig | Weak | Danish | CC | N | Major histocompatibility complex, class I, B | AS; HLAB; HLAC; SPDA1; HLA-B27; HLA-B73; MGC111087; HLA-B-7301; HLA-B | n/a | B15 | 272392 | Reinholdt J, Bay I, Svejgaard A. Association between HLA-antigens and periodontal disease. J Dent Res. 1977 Oct;56(10):1261-3. |
| HLA-B | CP | 40 | 120 | 0.0004 | Weak | South Indian | CC | N | Major histocompatibility complex, class I, B | AS; HLAB; HLAC; SPDA1; HLA-B27; HLA-B73; MGC111087; HLA-B-7301; HLA-B | 1 | B15 | 17209778 | Roshna T, Thomas R, Nandakumar K, Banerjee M. A case-control study on the association of human leukocyte antigen-A*9 and -B*15 alleles with generalized aggressive periodontitis in an Indian population. J Periodontol. 2006 Dec;77(12):1954-63. |
| HLA-B | CP | 25 | 50 | 0.0059 | Weak | Mixed | CC | E | Major histocompatibility complex, class I, B | AS; HLAB; HLAC; SPDA1; HLA-B27; HLA-B73; MGC111087; HLA-B-7301; HLA-B | n/a | B5 | 6584591 | Goteiner D, Goldman MJ. Human lymphocyte antigen haplotype and resistance to periodontitis. J Periodontol. 1984 Mar;55(3):155-8. |
| HLA-B | CP | 102 | 204 | 0.014 | Weak | Caucasian | CC | N | Major histocompatibility complex, class I, B | AS; HLAB; HLAC; SPDA1; HLA-B27; HLA-B73; MGC111087; HLA-B-7301; HLA-B | n/a | B*14 | 12296785 | Machulla HK, Stein J, Gautsch A, Langner J, Schaller HG, Reichert S. HLA-A, B, Cw, DRB1, DRB3/4/5, DQB1 in German patients suffering from rapidly progressive periodontitis (RPP) and adult periodontitis (AP). J Clin Periodontol. 2002 Jun;29(6):573-9. |
| HLA-B | CP | 102 | 204 | 0.014 | Weak | Caucasian | CC | G | Major histocompatibility complex, class I, B | AS; HLAB; HLAC; SPDA1; HLA-B27; HLA-B73; MGC111087; HLA-B-7301; HLA-B | n/a | B*14 | 12485327 | Reichert S, Stein J, Gautsch A, Schaller HG, Machulla HK. Gender differences in HLA phenotype frequencies found in German patients with generalized aggressive periodontitis and chronic periodontitis. Oral Microbiol Immunol. 2002 Dec;17(6):360-8. |
| HLA-B | CP | 102 | 204 | ns | None | Caucasian | CC | N | Major histocompatibility complex, class I, B | AS; HLAB; HLAC; SPDA1; HLA-B27; HLA-B73; MGC111087; HLA-B-7301; HLA-B | n/a | n/a | 12941076 | Stein J, Reichert S, Gautsch A, Machulla HK. Are there HLA combinations typical supporting for or making resistant against aggressive and/or chronic periodontitis? J Periodontal Res. 2003 Oct;38(5):508-17. |
| HLA-B.HLA-DQB1 | CP | 102 | 204 | ns | None | Caucasian | CC | N | n/a | n/a | n/a | Multiple Genes | 12941076 | Stein J, Reichert S, Gautsch A, Machulla HK. Are there HLA combinations typical supporting for or making resistant against aggressive and/or chronic periodontitis? J Periodontal Res. 2003 Oct;38(5):508-17. |
| HLA-B.HLA-DQB1 | CP | 102 | 204 | ns | None | Caucasian | CC | N | n/a | n/a | n/a | Multiple Genes | 12941076 | Stein J, Reichert S, Gautsch A, Machulla HK. Are there HLA combinations typical supporting for or making resistant against aggressive and/or chronic periodontitis? J Periodontal Res. 2003 Oct;38(5):508-17. |
| HLA-B.HLA-DRB1 | CP | 102 | 204 | 0.029 | Weak | Caucasian | CC | N | n/a | n/a | n/a | Multiple Genes | 12941076 | Stein J, Reichert S, Gautsch A, Machulla HK. Are there HLA combinations typical supporting for or making resistant against aggressive and/or chronic periodontitis? J Periodontal Res. 2003 Oct;38(5):508-17. |
| HLA-C | AgP | 18 | 18 | ns | None | Black | CC | N | Major histocompatibility complex, class I, C | D6S204; FLJ27082; HLA-Cw; HLA-Cw12; HLA-JY3; HLC-C; PSORS1; HLA-C | 4 | Multiple SNPs | 3457042 | Cogen RB, Roseman JM, Al-Joburi W, Louv WC, Acton RT, Barger BO, Go RC, Rasmussen RA. Host factors in juvenile periodontitis. J Dent Res. 1986 Mar;65(3):394-9. |
| HLA-C | AgP | 60 | 3791 | ns | None | Turkish | CC | N | Major histocompatibility complex, class I, C | D6S204; FLJ27082; HLA-Cw; HLA-Cw12; HLA-JY3; HLC-C; PSORS1; HLA-C | 4 | Multiple SNPs | 8811476 | Firatli E, Kantarci A, Cebeci I, Tanyeri H, Sönmez G, Carin M, Tuncer O. Association between HLA antigens and early onset periodontitis. J Clin Periodontol. 1996 Jun;23(6):563-6. |
| HLA-C | AgP | 50 | 152 | ns | None | Caucasian | CC | N | Major histocompatibility complex, class I, C | D6S204; FLJ27082; HLA-Cw; HLA-Cw12; HLA-JY3; HLC-C; PSORS1; HLA-C | n/a | n/a | 12296785 | Machulla HK, Stein J, Gautsch A, Langner J, Schaller HG, Reichert S. HLA-A, B, Cw, DRB1, DRB3/4/5, DQB1 in German patients suffering from rapidly progressive periodontitis (RPP) and adult periodontitis (AP). J Clin Periodontol. 2002 Jun;29(6):573-9. |
| HLA-C | AgP | 50 |
Stay updated, free dental videos. Join our Telegram channel

VIDEdental - Online dental courses


