Chapter 4
Efficacy of Root Canal Disinfection
Ashraf F. Fouad
Department of Endodontics, Prosthodontics and Operative Dentistry, University of Maryland, Baltimore, MD, USA
Goals of endodontic treatment and of root canal disinfection
Root canal disinfection, rather than mere debridement, has become a central goal of endodontic treatment in recent years. This approach developed from the realization that microbial infection is the principal cause for the initiation and persistence of endodontic disease (1, 2). Microbial presence and persistence following treatment is also likely the reason for the differences in clinical outcomes between cases with preoperative apical lesions and cases without lesions (3, 4). It has become evident from the examination of longitudinal outcome studies that this distinction in outcomes between vital cases and cases with preoperative lesions is present despite the fact that many of the cases with vital pulps in these studies are treated by trainees who have less clinical experience (3, 5). Root canal systems in cases with vital pulp are likely to have residual debris in the root canal following the completion of instrumentation, regardless of the instrumentation technique; yet the long-term success remains high. Likewise, retreatment cases without preoperative apical lesions also have a much better outcome compared to those with apical lesions (3, 6), despite numerous studies showing the persistence of debris following the instrumentation of a previously treated case. Thus it is not the debris that is associated with treatment outcome, but rather whether or not there is evidence that the cases involved may have had bacterial biofilms preoperatively. Treatment procedures in these cases may not have completely eliminated the bacterial biofilm in a large number of cases, and despite obturation that may be adequate, the lesions persisted. In fact, a well-performed animal study has documented this relationship between residual bacteria and lesion persistence, by showing that the quality of obturation was an important factor only if bacteria persisted at the time of obturation (2). Another animal study corroborated these findings by showing that instrumented, obturated, and sealed root canals in teeth with apical lesions had similar histological evidence of healing at 6 months to teeth that had instrumentation and coronal seal but without obturation (7).
Clinical outcome studies have documented the association between the presence of intracanal bacteria and the presence and persistence of apical disease. An association may or may not involve a causal relationship. In this case, causation has been demonstrated by several animal studies. These studies have shown the causal relationship between bacteria (8, 9) or bacterial cell wall components (10–12), and the induction and magnitude of apical disease. There have also been other animal studies (2) and clinical studies (13–16) showing improved apical healing in cases in which clinical intervention has resulted in the elimination of cultivable bacteria, although this has not been a universal finding (17).
Given these findings, it could be concluded that one of the main aims of root canal treatments is to provide a bacteria-free space that can be effectively obturated, which will likely improve the outcomes of cases with preexisting apical lesion. However, the root canal space is generally too complex anatomically to allow the clinician to achieve this aim using clinical techniques. In addition, there are areas in the root canal environment, such as surface irregularities, dentinal tubules, and lateral canals, that may become infected, but have a hitherto unknown effect on treatment outcome (18). Finally, the clinician does not have a clinical metric that allows him/her to determine whether root canal disinfection has been adequately accomplished.
Detection of microbial pathogens
In order to adequately discuss the possibility of microbial elimination, one has to have a good understanding of the methods available for microbial detection (Table 4.1). Microbial elimination should be analyzed in the context of how the microorganisms are detected. As different methods of microbial detection have different specificities and sensitivities, it follows that these different methods provide differing thresholds for microbial detection.
Table 4.1 Common methods of detection of root canal microorganisms.
| Detection method | Description | Limitations |
| Aerobic and anaerobic culturing | Requires various generic and specific media for specific microorganisms | Many bacteria are not cultivable, or may be rendered viable but uncultivable by temporary stressors in the system |
| PCR with species-level primers | Enables very specific identification of a particular species | Limited to one species, may detect the DNA of the dead organism |
| PCR with genus-level primers | Allows identification of a genus, may be followed by direct sequencing of the product to determine the species | Multiple species in the same genus may be present in the specimen, rendering the distinction among them difficult |
| PCR with broad-range 16S rDNA primers | Identifies the presence or absence of any bacteria, may be followed by cloning and sequencing to identify species | Laborious and expensive; only a limited number of clones could be identified from each sample |
| DNA–DNA hybridization with molecular probes | Allows identification of selected species or all bacteria | Lacks amplification; only a limited number of species is detectable using this method |
| Fluorescent in situ hybridization | Detects bacteria (or fungi) histologically in their undisturbed sites | Can detect bacteria but not differentiate a lot of individual species |
| Detection of messenger RNA | Allows detection of live bacteria | Technically difficult and only a limited number of targets available for detection |
| Proteomics | Allows functional detection of bacterial or fungal proteins, which demonstrate their activities | Complex analysis and differentiation of host and bacterial proteins |
| Monoclonal antibodies against certain microorganisms | Detects specifically certain microorganisms histologically in tissues | Antibodies may cross-react with other species |
| Scanning electron micrographs | Shows biofilms and their relationship to substrate | Usable only for hard tissues; industrial SEM needed to avoid distortion of tissues |
| Transmission electron micrographs | Shows bacteria and fungi in situ at very high resolution | Very laborious to perform, particularly for larger specimens |
| Brown and Brenn histologic bacterial staining | Stains bacteria in situ | Cannot stain gram-negative bacteria, and processing of specimens may eliminate many bacteria |
| BacLight live/dead stains | Stains vital and dead bacteria | Limited to smears or histologic sections |
| Bacterial luminescence | Uses bacterial species with light emitting characteristics that can be measured in a gradient | Requires use of special dark chamber for minimal light detection |
| Pyrosequencing | Allows depth of coverage to identify the most amount of taxa of any technique | Does not have resolution to determine individual species with great precision |
Endodontic infections are polymicrobial in nature, and are a result of microbial biofilms that populate the carious lesion causing pulpal inflammation. Following pulpal necrosis, there is microbial transformation and growth in the root canal environment that result in apical disease. These microbial biofilms are primarily bacterial in nature. However, archeal (19–21) and fungal species also have been identified in endodontic infections (22, 23), and viral infections appear to exacerbate apical disease (24–27). Other forms of microbial disease, such as from eukaryotic parasites, have not been described in endodontic infections. As the overwhelming evidence for the causation and persistence of endodontic disease is related to bacterial pathogens, this domain of microorganisms remains the principal target of microbial analyses as well as antimicrobial therapy. Bacterial species in root canal infections appear to be derived from those that populate deep carious lesions, the gingival sulcus, and the rest of the oral cavity (28).
Since 19th century (29), cultivation of bacterial cells has been performed in the majority of studies that have linked bacteria to the initiation of disease or to the healing following treatment. It has been known ever since that what can be cultured in the laboratory is a small fraction of what can be seen in a smear from the root canal under the microscope. Therefore, investigators have always sought to improve bacterial cultivation techniques. In the 1970s, newer cultivation methodologies that involved the use of anaerobic environment have shown that most root canal bacterial species are facultative or strict anaerobes (30–32). Therefore, many studies continue to employ anaerobic cultivation procedures, together with special enriched culture media, in order to maximize the cultivation of bacteria obtained from the root canal environment. However, the advent of molecular studies in the past decade has provided technologies that allow the efficient analyses of much more bacterial species, at a lower amount in the sample (Table 4.1).
These molecular methods rely on the detection of microbial DNA (33), RNA (34), or proteins (35). The detection of microbial DNA depends primarily on the identification of sequences in the 16S rRNA gene (which is uniquely present in bacterial and archeal species (19), other genes in bacteria (36), or the 18S rRNA gene in fungi (37). The analysis of these genes allows the investigators to search public databases that contain tens of thousands of the respective microbial species and thus perform a more comprehensive analysis of microbial presence. DNA-based molecular procedures generally involve either hybridization methods for identification of DNA signature sequences for specific bacteria (38) or amplification methods that employ the polymerase chain reaction (PCR) technology (33, 39). Amplification, using PCR-based methods, allows the detection of very small amounts of target organisms, thereby remarkably increasing the sensitivity of the test. Depending on the specificity of the primers used in PCR, the test may be used for the detection of specific species, several species in a genus, or for analyzing a large array of microorganisms present in the sample (33, 36, 39). The last method usually employs amplification of a large segment of the 16S rRNA gene that includes conserved regions of the gene thereby amplifying the genes from many different bacterial taxa. This is then followed by cloning the amplified genes into a commercially available vector (such as the ones made in Escherichia coli). Allowing the E. coli cells to grow then provides colonies of the different bacterial clones amplified. The 16 S rRNA gene from each of these colonies can then be sequenced to determine the identity of the bacteria in the original sample from which it was cloned. This process is referred to as cloning and sequencing with broad-range primers (40–42).
Contemporary analyses of human microbiome (all bacteria and fungi that populate the human body) revealed that only about 10% of them can be cultivated (43). Current procedures rely on advanced in silico sequencing methodologies that avoid the inefficient biological cloning methods, and simultaneously analyze tens of thousands of sequences. This is known as pyrosequencing and has revealed a wide diversity of endodontic bacterial taxa, which could not be appreciated from cultivation or even earlier clonal analysis studies (44, 45).
Analyses of functional genes other than the 16S rRNA gene can also be performed, especially to identify certain traits in the specimen, such as virulence factors (46–48) or antibiotic resistance genes (49, 50). However, given the enormity of bacterial genomes and reduced cost of massive sequencing, contemporary studies employ the “shot gun” sequencing approaches to perform the so-called metagenomic analyses of a polymicrobial specimen. This could provide a wealth of information about the virulence of the microorganisms present, the presence of antibiotic resistance genes as well as the host response (51). However, metagenomic analyses have not been extensively performed in endodontic infections. DNA analysis in general, whether for identification or metagenomic analyses, has some limitations despite enabling an extensive examination of microbial presence. These include the fact that DNA may survive after bacterial cells die (52, 53), and that many of the genes present in a specimen may not actually be expressed into RNA and translated into proteins.
Analysis of RNA provides information on genes that have become expressed in the environment and thus can be considered a functional analysis of the specimen. Detection of messenger RNA also provides evidence for the viability of cells, because RNA degrades rapidly after the bacterial or fungal cell from which it is detected dies. Only a limited number of studies have been performed using this method in the endodontic literature (34). Proteomic analysis provides perhaps the most direct evidence for specific microbial activities and virulence, and how the proteins present compare to those of the host. Proteomic analysis is also considered a functional analysis of the specimen and has not been sufficiently utilized in analyzing endodontic infections (35).
With respect to the detection of viable bacterial cells, another method that has been employed frequently is by using the vital stains, most notably the BacLight system (54). This method involves the study of two stains of bacterial cells, of which Syto 9 stains both vital and dead bacteria whereas propidium iodide stains dead cells only. The staining can be performed of smears from the site to be studied, or of histological sections, to observe the cells in situ. The imaging is usually performed with a confocal laser scanning microscope, which can visualize the fluorescence of the stains at the appropriate wavelength for each of them. This allows the visualization of viable cells, which will be the ones that stain with Syto 9 but not propidium iodide. In endodontics, this method has been used to demonstrate that calcium hydroxide may render bacterial cells viable but not cultivable (54, 55), and that the biofilm status provides a mechanism for bacterial cells to resist stress from increased alkalinity (56).
Staining of bacteria in situ using histobacteriological methods has long been a powerful method to demonstrate the presence and location of bacterial cells. This has traditionally been accomplished using the Brown and Brenn, modified gram staining, method (57–59), or transmission electron microscopy (60, 61).
The observation of bacterial cells in situ has also been performed using molecular probes that hybridize with bacterial signatures in the bacterial genome and are attached to fluorescent markers. This method commonly referred to as fluorescent in situ hybridization (FISH) has been used to provide elegant staining of bacteria within persistent apical lesions (62). This study demonstrated how this method could identify bacteria within the center of the surgical specimens of apical lesions, without any concerns about the contamination from gingival and oral sources on the periphery of the enucleated lesions.
Histological processing of tissues involves fixation with potent antimicrobials such as formaldehyde or alcohol, followed by decalcification of mineralized tissues in strong acids, or extended soaking in ethylenediaminetetraacetic acid (EDTA), and continued by paraffin embedding, sectioning, and staining, and so on. All these steps may lead to the killing and removal of any bacteria present in the tissues. An ideal in situ observation method is one that does not involve the destruction and processing of tissues. An ideal example of the use of bioluminescent bacterial strains in intact teeth to demonstrate the efficacy of root canal preparation and irrigation depth has been presented in Figure 4.1 (63–65).
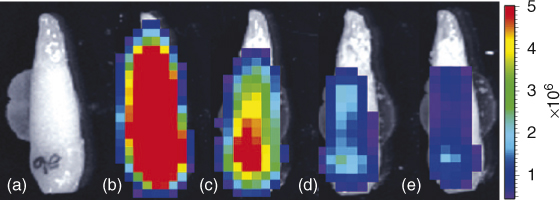
Figure 4.1 Images of representative tooth. (a) Background, no bacteria, (b) 1 × 106 Pseudomonas fluorescens 5RL in root canal. Bacterial cell count was determined by culturing the same volume of inoculum. (c–e) After 6 ml irrigation delivered using 28 gauge Max-I-Probe needle positioned 1 mm from WL after root canal instrumented to (c) size 36, (d) size 60, and (e) size 77. Color bar on right gives bioluminescence image units (photons/s/cm2/sr).
(Falk and Sedgley (63). Reproduced with permission of Elsevier.)
As noted before, several studies have been performed on the association of cultivated bacteria at the time of root canal obturation and long-term outcomes of treatment. Most of these studies have shown a direct correlation between microbial elimination and healing of apical lesions; however, this conclusion is not unequivocal. Therefore, at the time of writing, it is not known whether the lack of universal predictive nature of cultivation results on treatment outcomes is due to the lack of sensitivity of the technique, its inability to detect many microbial taxa, inability to predict whether the residual microflora will survive and have access to apical tissues, or due to changes that occur with time to root canal microflora following obturation. Very few studies have done these analyses using molecular methods. One animal study referred to earlier has shown that microbial presence at the time of follow-up (which was 2–2.5 years after treatment) showed a much higher association with apical healing than microbial presence at the time of obturation (2).
Sampling as a source of bias
In order to detect microbial presence in root canals, a sample has to be obtained and analyzed, as stated before. The tooth surface involved is typically treated by 30% hydrogen peroxide and disinfected by 5% tincture of iodine (66), with or without 5.25–6% sodium hypochlorite (67). The halides are then inactivated with 5% sodium thiosulfate, so as not to kill root canal bacteria. If the tooth has caries or a defective restoration, this sequence of solutions is repeated after the caries and restoration are removed, but before the canal is entered. A mi/>
Stay updated, free dental videos. Join our Telegram channel

VIDEdental - Online dental courses


