Chapter 3
Biofilms in Root Canal Infections
Christine M. Sedgley
Department of Endodontology, Oregon Health and Science University, Portland, OR, USA
Rogério de Castilho Jacinto
Department of Semiology and Clinics, Federal University of Pelotas, Pelotas, RS, Brazil
Introduction
Biofilms are microbial communities composed of microcolonies that contain cells irreversibly attached to a substratum, an interface, or each other, as opposed to being suspended in liquid (planktonic mode). Biofilm formation has been described as an ancient prokaryotic adaptation allowing survival in hostile environments (1–3). The process encompasses attachment of microbial cells to a surface, followed by cell proliferation, adherence to other bacteria, production of a matrix, and microcolony maturation (4). Dispersal of cells allows the formation of new biofilm microcolonies (5). A key characteristic of biofilms is their highly heterogeneous composition that can vary greatly under different environmental conditions. Of particular clinical significance is that microbial cells in biofilms can utilize a variety of mechanisms that enable survival under challenging conditions (6–10). For example, biofilms have been regarded as the primary causative agent for many hospital-acquired infections as well as persistent and chronic infections such as cystic fibrosis pneumonia (4, 8).
Seminal studies published several decades ago established the role of microorganisms as the primary etiologic agents of apical infections (11–14). In these and many subsequent evaluations of the root canal (RC) microflora, strains were typically grown as single-species, planktonic cultures. More recently, it has been recognized that microorganisms exist in the RC system as multispecies biofilm communities (15–20). This chapter describes the general characteristics of biofilms and analyzes biofilms associated with RC infections.
General characteristics of biofilms
Components of biofilms
Microbial cells occupy only a small portion of the biofilm. The majority of the biofilm structure is a highly heterogeneous matrix composed of extracellular polymeric substances (EPS) produced by cells within the biofilm. The EPS matrix can account for greater than 90% of the dry mass of most biofilms and provides multiple functions. These include provision of a scaffold for the biofilm, water retention, protection, an energy sink, ionic exchange, sorption of organic and inorganic compounds, nutrient source, and exchange of genetic information (21). From a clinical perspective, the EPS can act as a physical barrier to antimicrobial agents such as antibiotics and disinfectants (21, 22). The major component of the EPS matrix is water, with channels throughout the biofilm structure facilitating the inflow of nutrients and the outflow of waste materials (23, 24). Other important components of the highly hydrated EPS include extracellular polysaccharides, proteins, and DNA (eDNA, extracellular deoxyribonucleic acid), as well as other elements (5, 21) (Figure 3.1).
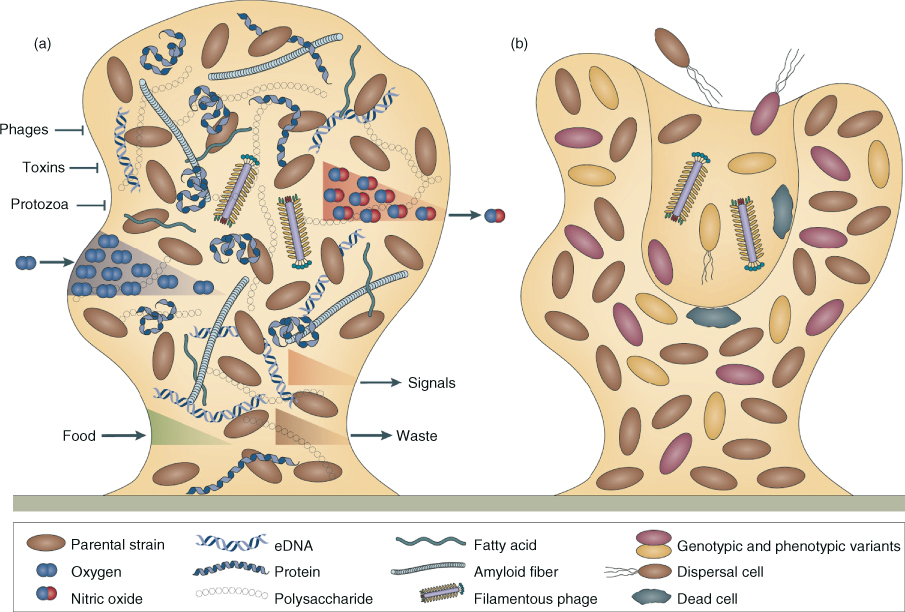
Figure 3.1 Components of biofilms. (a) Microcolonies in the mature biofilm are characterized by an extracellular polymeric substances (EPS) matrix, composed of polysaccharides, proteins, extracellular DNA (eDNA), and other elements. The EPS matrix functions as a shield to protect the bacterial community or population from predators such as protozoa or lytic phages, as well as from chemical toxins (e.g., biocides and antibiotics). The EPS matrix may help to sequester nutrients and, along with the underlying microorganisms, is also responsible for the establishment of gradients (e.g., oxygen and nutrients diffusing inward, and waste products as well as signals such as nitric oxide diffusing outward); (b) at the time of dispersal, microcolonies undergo cell death and lysis along with active dispersal of motile microorganisms to leave behind hollow colonies.
(Reprinted by permission from Macmillan Publishers Ltd. from: Nature Reviews in Microbiology (McDougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S.)
Extracellular polysaccharides provide a scaffold for proteins mediating adhesion of the biofilm to the surface and intercellular attachments (22, 25). They can also contribute to antimicrobial resistance by preventing antibiotics from reaching their site of action (26) and by protecting the biofilm from the host defenses (27). Extracellular proteins supply the essential structural and enzymatic functions. For example, glucan-binding proteins produced by Streptococcus mutans provide a link to exopolysaccharides (28). Enzymes within the EPS matrix provide carbon and energy sources by digesting polymers (21) and degrade the EPS matrix to facilitate dispersion of microbial cells that can form new biofilms (29). eDNA is involved in the adhesion, aggregation, cohesion, and exchange of genetic information (21, 30–32). eDNA also plays a critical role in the initial establishment of biofilms (32) (Figure 3.2).

Figure 3.2 Ultrastructural analysis of eDNA during the initial establishment of E. faecalis biofilms. (a, b) Immuno-SEM micrographs demonstrating localization of eDNA near the septum in early (4 h) E. faecalis biofilms; (c) endogenous lyses of cells in an older (48 h) biofilm display an entirely different morphology from that seen in early biofilms, as DNA (asterisks) is released from a ruptured cell (arrow). Bars, 500 nm.
(Barnes et al. (32). Reproduced with permission of Gary Dunny.)
Cell–cell communication in biofilms
Quorum sensing
The distance between microorganisms and their spatial distribution within biofilms are critical factors for intermicrobial communication processes in biofilms (1). Certain bacteria communicate and coordinate behavior via the constitutive synthesis of small signaling molecules using an intercellular signaling system called quorum sensing (33, 34) (Figure 3.3). Quorum sensing allows the coordinated regulation of the expression of key proteins in biofilms so that when a high density population reaches a certain threshold (“quorum”), the concentration of normally low levels of certain signal molecules becomes high enough to act as autoinducers (AIs) that trigger a synchronized response throughout the biofilm population (35, 36). AI-2 signal is used by both gram-negative and gram-positive bacteria. Gram-negative bacteria also use N-acyl homoserine lactone-based signaling while gram-positive bacteria utilize small peptides (33, 37–39).
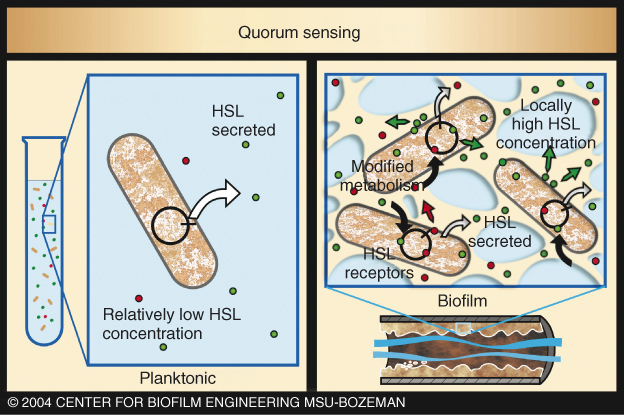
Figure 3.3 Quorum sensing in biofilm and planktonic conditions. Quorum sensing occurs when the concentration of normally low levels of certain signal molecules (e.g., homoserine lactones, or HSLs) becomes high enough to act as autoinducers (AIs) that trigger a synchronized response throughout the biofilm population. As biofilm cells are held together in dense populations, the secreted HSLs attain higher concentrations. HSL molecules then recross the cell membranes and trigger changes in genetic activity.
(Reproduced with permission of the Center for Biofilm Engineering, Montana State University.)
Horizontal gene transfer (HGT)
Interactions between genetically distinct bacteria are involved in the establishment and maintenance of biofilms (1) and can provide an accommodating environment for the transfer of genetic material (40). Horizontal gene transfer (HGT) occurs by three basic methods: transformation, conjugation, and transduction, and allows the movement of genetic information both within and between species via plasmids, bacteriophages, and transposons (Figure 3.4).
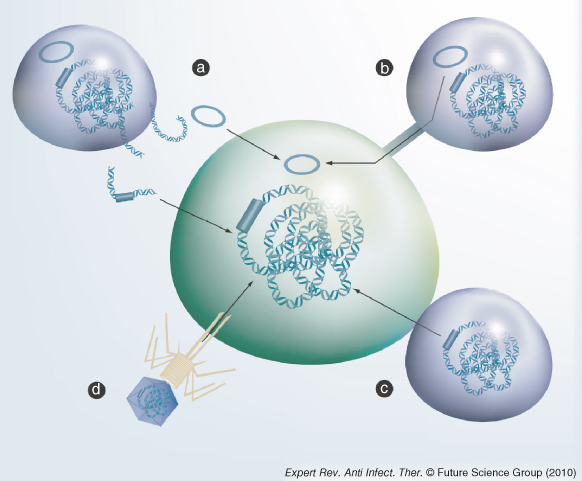
Figure 3.4 Horizontal gene transfer of DNA. The bacterial cells are represented by the green and purple ovals. The DNA is represented by the helix. Transposons and plasmids are either blue bars or circles. The arrows show the direction of transfer of the DNA. (a) Transformation: the donor cell (top left) has lysed and the DNA released into the environment. This can be taken up by competent bacteria and incorporated into the recipient genome; (b) conjugation of plasmids through a pilus; (c) conjugation of transposons via a mating pore; (d) transduction mediated by the injection of DNA by a bacteriophage.
(Reprinted with permission from Roberts AP, Mullany P. Oral biofilms: a reservoir of transferable, bacterial, antimicrobial resistance. Expert Review of Anti-Infective Therapy 8(12):1441–1450. Expert Reviews Ltd © 2010.)
Biofilm growth has been shown to enhance genetic exchange via transformation, as demonstrated in several streptococcal species naturally competent for transformation (42, 43). However, the most efficient HGT process in bacteria is conjugation, with the requirement for cell-to-cell contact distinguishing conjugation from transduction and transformation (Figure 3.4). The ability of plasmids to conjugatively transfer between species has been demonstrated in oral streptococci (44–46), and between Treponema denticola and Streptococcus gordonii in experimental biofilms (47). In the endodontic microflora, several enterococcal isolates recovered from patients in Sweden exhibited a characteristic response to enterococcal-derived pheromone that supported the possibility of highly efficient pheromone-induced conjugative transfer of genetic elements in RC infections (48). This premise was subsequently supported in an ex-vivo model through the demonstration of bidirectional transfer of a conjugative plasmid in RCs occurring between S. gordonii and Enterococcus faecalis (49) (Figure 3.5). Taken together, these findings suggest the feasibility of RC strains containing conjugative plasmids with genes that enhance survival to transfer these properties to other strains in the RC biofilm.
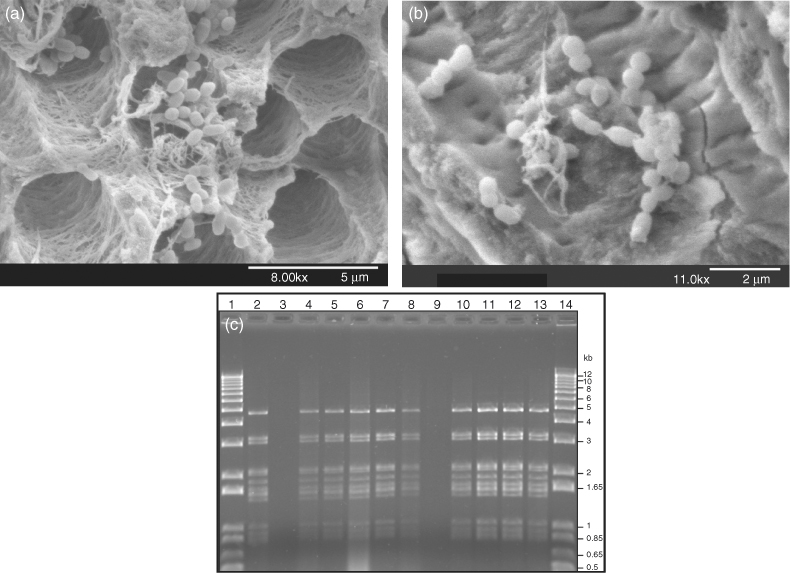
Figure 3.5 Horizontal gene transfer (HGT) in the root canal. Scanning electron micrographs showing accumulations of E. faecalis JH2-2/pAM81 and S. gordonii Challis-Sm (a) 24 h and (b) 72 h after inoculation into the root canal. Bidirectional HGT of the plasmid pAM81 was confirmed by the purification of the plasmid in transconjugants; (c) confirmation of conjugative transfer of pAM81 from E. faecalis to S. gordonii (lanes 2–7) and from S. gordonii to E. faecalis (lanes 8–13): pAM81 plasmid DNA digested with HindIII in donor strains (lanes 2 and 8) that were transferred to plasmid-free recipient strains (lanes 3 and 9) that resulted in transconjugant strains with pAM81 (lanes 4–7 and 10–13). Lanes 1 and 14 show molecular size marker.
(Reprinted from Journal of Endodontics, 34(5), Sedgley CM, Lee EM, Martin MJ, Flannagan SE. Antibiotic resistance gene transfer between Streptococcus gordonii and Enterococcus faecalis in root canals of teeth ex-vivo, 570–574 © 2008, with permission from Elsevier.)
Responses to stress and environmental challenges
In the ever-changing biofilm environment, microbial cells encounter a multitude of stresses and challenges. These include exposure to nutrient limitation, reactive oxygen and nitrogen species, membrane damage, and elevated temperature (10). Exposure to antimicrobial agents also places biofilm communities under stress.
A diverse range of global stress responses by cells exposed to these challenges can facilitate their survival (50). For example, global response systems (GRS) such as the alternative sigma factors RpoS (51) and RpoH (52), gene repressor LexA (53), and small molecule effectors, such as (p)ppGpp that induce the stringent response (54), act by modulating intracellular metabolic processes to enable adaptation and survival. Resultant stress responses include downregulation of error-correcting enzymes, upregulation and activation of error-prone DNA polymerases, and HGT of mobile genetic elements (55, 56). Two-component toxin–antitoxin (TA) systems are also involved in the responses to stress stimuli; the toxins are stable proteins directed against specific intracellular targets, and the antitoxins are degradable proteins or small RNAs that neutralize the toxin or inhibit toxin synthesis (57). TA systems are involved in persister cell formation (58) and quorum sensing (59) and may regulate the switch from planktonic to biofilm lifestyles (60).
As a direct consequence of stresses and the resulting responses, the availability of nutrients and electron acceptors can vary considerably throughout the biofilm, and the growth stage of microbial cells can range from rapidly growing to dormant. This results in variations in metabolic activity, gene expression patterns, and potential phenotypic variations throughout the biofilm community (7). Cells might adapt by turning certain genes on or off or by random gene switching, and fitter mutants might arise as part of a natural selection process (Figure 3.6).
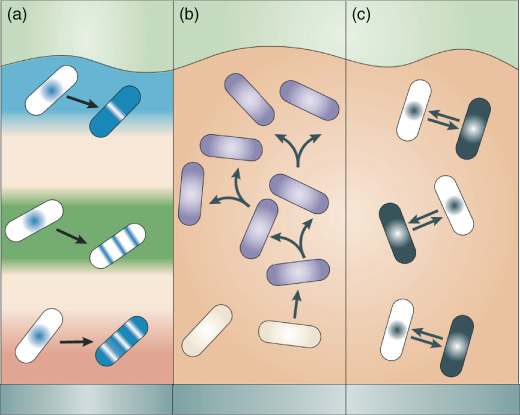
Figure 3.6 Multiplicity of phenotypic states in biofilms. Three hypothesized mechanisms of phenotypic diversification in a biofilm. (a) Physiological adaptation. Cells respond adaptively to local environmental conditions by turning on or off certain genes; the responses depend on the local chemical microenvironment and, therefore, allow for a range of distinct localized adaptations; (b) genotypic variation and natural selection. A mutation or chromosomal rearrangement results in a variant (purple) that multiplies according to its fitness in the biofilm; (c) stochastic gene switching. Cells toggle between discrete physiological states by gene switching, which is random in nature.
(Reprinted by permission from Macmillan Publishers Ltd from: Nature Reviews (Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol 6(3):199–210) © 2008.)
Responses to antimicrobials in biofilms
Microorganisms in biofilm communities have several options available to resist the detrimental effects of antimicrobial therapy (4, 61, 62) (Figure 3.7). For example, the thickness of the biofilm, as well as the concentration and ability of the antimicrobial to penetrate the EPS matrix, can determine the extent of exposure of cells in biofilms. Thus, while the superficial cells of the biofilm community are rapidly exposed to high concentrations of antimicrobials, exposure of deeper parts depends on the ability of the antimicrobial to diffuse through the biofilm matrix (61). The EPS matrix can slow down the diffusion of the agent (63) but does not necessarily prevent diffusion (64). Multidrug resistance efflux pumps are also considered to contribute to antibiotic resistance in biofilms (65–67).
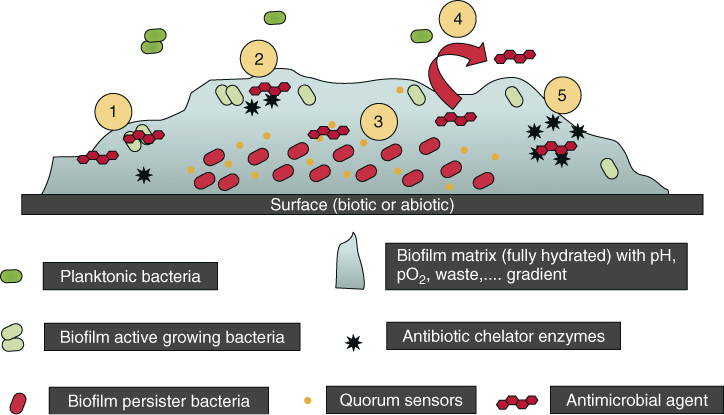
Figure 3.7 Some proposed biofilms associated with resistance mechanisms: (1) antimicrobial agents may fail to penetrate beyond the surface layers of the biofilm. Outer layers of biofilm cells absorb damage. Antimicrobial agents’ action may be impaired in areas of waste accumulation or altered environment (pH, pCO2, pO2, etc.); (2) antimicrobial agents may be trapped and destroyed by enzymes in the biofilm matrix; (3) altered growth rate inside the biofilm. Antimicrobial agents may not be active against nongrowing microorganisms (persister cells); (4) expression of biofilm-specific resistance genes (e.g., efflux pumps); (5) stress response to hostile environmental conditions (e.g., leading to an overexpression of antimicrobial agent-destroying enzymes).
(Reprinted by permission from Macmillan Publishers Ltd: Clinical Pharmacology and Therapeutics (del Pozo JL, Patel R. The challenge of treating biofilm-associated bacterial infections. Clin Pharmacol Ther. 2007 82(2):204–209) © 2007.)
Oxygen levels and metabolic rates are reduced at the center of a microcolony compared with near its surface (68, 69). This can result in localized limited or absent metabolic activity of microbial cells deeper in the biofilm as well as induce cells to enter stationary phase and thus lose susceptibility to killing by antibiotics that target dividing cells (70). Persister cells have been associated with recalcitrant biofilm infections (9, 71). Persister cells are phenotypic variants that are genetically identical to the susceptible cells within a clonal population (72, 73). On exposure of the biofilm to antibiotics, persister cells demonstrate very slow or arrested growth (“dormancy”) and diminished protein synthesis (9, 74). Following the treatment of biofilms with antibiotics such as β-lactams, the rapidly growing cells are killed, leaving the dormant cells to repopulate the biofilm. The formation of persister cells is thought to be through a combination of continuously occurring random events (“stochastic”), or in response to an environmental stimulus (“deterministic”) (9, 75).
Resistance to antimicrobial treatment is significantly higher in multispecies biofilms compared to single-species biofilms (76–78). Dual-species biofilms of S. mutans and Veillonella parvula were less susceptible to various antimicrobial agents (chlorhexidine, cetylpyrimidinium chloride, zinc chloride, erythromycin, hydrogen peroxide, and amine chloride) than single-species biofilms of the same microorganisms (76, 79). This may be attributable to the cooperative behavior of the community that enables survival on exposure to the antimicrobial agent (80), and potential advantages to be gained by residing in multispecies biofilms (77).
Root canal biofilms
Evaluation of root canal biofilms
The anatomy of the RC system is intricate and complex. In addition to the main canal(s), there can be lateral canals, accessory canals, apical ramifications, and isthmuses (81). All anatomical areas of the infected RC system have been shown to harbor bacterial cells organized as archetypal biofilm structures (82, 83). In 1987, Nair (15) described infected RCs containing clusters of “self-aggregating” colonies of one distinct type or “coaggregating” communities of several types that would now be described as biofilm structures. Since then, observations of RC biofilms in infected extracted teeth have utilized light microscopy (15, 17, 20, 82, 84), scanning electron microscopy (SEM) (85, 86), environmental SEM (87), transmission electron microscopy (TEM) (18, 88), confocal laser scanning microscopy (CLSM) and fluorescent in situ hybridization (FISH) (89), and various combinations of techniques (19, 90) (Figure 3.8). Collectively, these observational studies have shown that RC biofilms are highly variable.
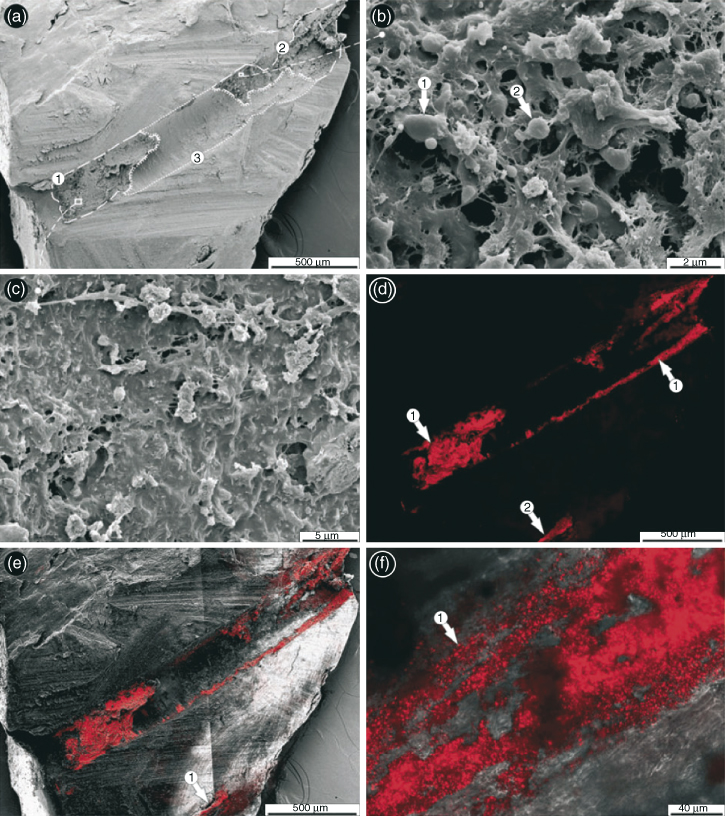
Figure 3.8 Imaging of endodontic biofilms by combined microscopy (FISH/CLSM–SEM). (a) Overview of split root canal. Certain parts of the canal surface were covered with a thick matrix layer (encircled areas 1, 2) whereas other regions showed only sparse and comparatively thin islands of matrix (encircled area 3); (b) the size and shape of some structures of the matrix suggested the presence of bacteria (arrows 1, 2), but without conclusive evidence; (c) other areas of the matrix consisted of so densely composed material that no traces of bacterial presence could be found; (d) After labeling the split tooth with the EUB338 (Cy3) probe, distinct parts of the canal showed a strong red fluorescence signal in the CLSM (arrows 1 and 2); (e) overlay of corresponding SEM and FISH/CLSM (a and d) images of the same regions revealed matching areas of the FISH signal and the amorphous matrix, suggesting the presence of bacterial biofilm. The laterally located FISH signal was within a lateral canal in the root (arrow 1); (f) higher magnifications of FISH-labeled root canal surface with CLSM indicated a biofilm composed of bacteria with short rod or coccus-like morphology (arrow 1).
(Reprinted by permission from John Wiley and Sons from Schaudinn C, Carr G, Gorur A, Jaramillo D, Costerton JW, Webster P. Imaging of endodontic biofilms by combined microscopy (FISH/cLSM – SEM). Journal of Microscopy 235(2):124–127 © 2009.)
Much effort has gone into the identification of the microbial component of clinical RC infections. Genera cultured from symptomatic and asymptomatic RC infections and aspirates from apical abscesses include Prevotella, Porphyromonas, Fusobacterium, Peptostreptococcus, Streptococcus, Lactobacillus, Enterococcus, Actinomyces, Propriobacterium, and Candida (12, 13, 91, 92). Compared to culture methods, molecular analyses of RC samples have shown a substantially more diverse microflora that is composed of a large proportion of unidentifiable and unculturable species (93, 94). Recent analyses using pyrosequencing methods have revealed an even greater microbial diversity (95–98) (Figure 3.9).
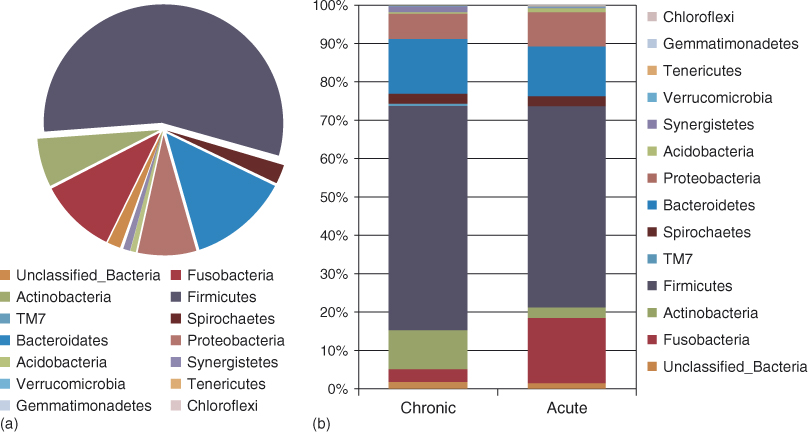
Figure 3.9 Relative abundance of different bacterial phyla in acute and chronic dental root canal infections. (a) Overall data; (b) data according to the clinical condition. Phylogenetic classification was based on Ribosomal Database Project Classifier analyses.
(Santos et al. (98). Reproduced from PLoS ONE 6(11): e28088. doi:10.1371/journal.pone.0028088.)
The recovery and analyses of the EPS matrix component of RC biofilms during clinical procedures are yet to be described. Considerable challenges exist in obtaining intact RC biofilm samples in a nondestructive manner. From a practical aspect, it is likely that paper points absorb planktonic microorganisms disrupted from the biofilm surface but are less likely to retrieve microorganisms located in the deeper parts of the biofilm or within the dentinal tubules. Similarly, file shavings are also unlikely to provide samples of “intact” RC biofilms. Sampling previously treated RCs poses further obstacles, particularly if the prior restoration and root filling material are difficult to remove.
Primary root canal infections
Many studies have confirmed that infected RCs harbor a multispecies population of facultative and strict anaerobic gram-positive and gram-negative bacteria, spirochetes, yeasts, archaea, and other unidentified species (95–103). In addition, Epstein–Barr virus may be associated with irreversible pulpitis and apical periodontitis (104), and papilloma virus and human herpes virus have been found in exudates from acute apical abscesses (AAAs) (105).
The microflora of carious dentin implicated in endodontic infections subsequent to pulpitis includes significant numbers of lactobacilli (106) and gram-negative bacteria (107). Both innate and adaptive immune responses modulate the relationship between tissue damage and the microflora in advancing caries (108). Many microorganisms cultured from endodontic infections have also been identified as commensals in the oral cavity. The transition from oral “commensal” to RC “pathogen” may reflect an innate ability to switch on genes that encode virulence factors (Figure 3.10) as well as other factors enabling survival and propagation in a different environment.
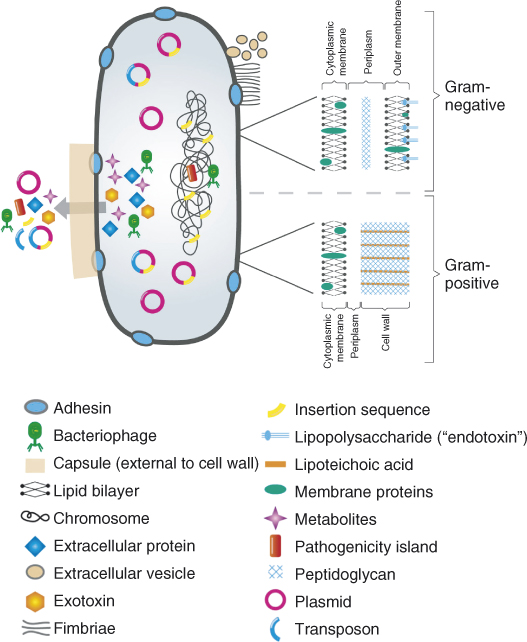
Figure 3.10 Virulence factors and associated genetic elements in bacteria.
(From Sedgley CM. Virulence of endodontic bacterial pathogens. Chapter 7;130-51. Endodontic Microbiology. Editor, Fouad AF. Wiley-Blackwell Publishing, Ames, IA. This material is reproduced with permission of John Wiley & Sons, Inc. © 2009.)
Bridging, or coaggregation, between two or more bacterial species is an important process in multispecies biofilm formation (1). In dental plaque, Fusobacterium nucleatum is considered to be a strategic microorganism as evidenced by its ability to coaggregate with numerous other species and mediate adherence of early and late colonizers (110). Similarly, in infected RCs, the commonly recovered Fusobacterium species appears to participate in synergistic interactions in RC infections (111). F. nucleatum exhibits strong associations with Peptostreptococcus micros, Porphyromonas endodontalis, and Campylobacter rectus (112, 113) and has been shown to enhance the pathogenicity of Porphyromonas gingivalis in mouse models (114). In addition, coaggregation assays have shown that fusobacteria form coaggregates with many species including Prevotella, Streptococcus (115), E. faecalis, Streptococcus anginosus, Peptostreptococcus anaerobius, and Prevotella oralis (116) (Figure 3.11). Other positive associations shown to occur in RC strains are between Prevotella intermedia and P. micros, and between P. anaerobius and Eubacteria and P. anaerobius (113). In addition, in guinea pigs, Parvimonas micra enhanced the pathogenicity of Bacteroides melaninogenicus (Prevotella melaninogenica) and Bacteroides asaccharolyticus (Porphyromonas spp.) (117).
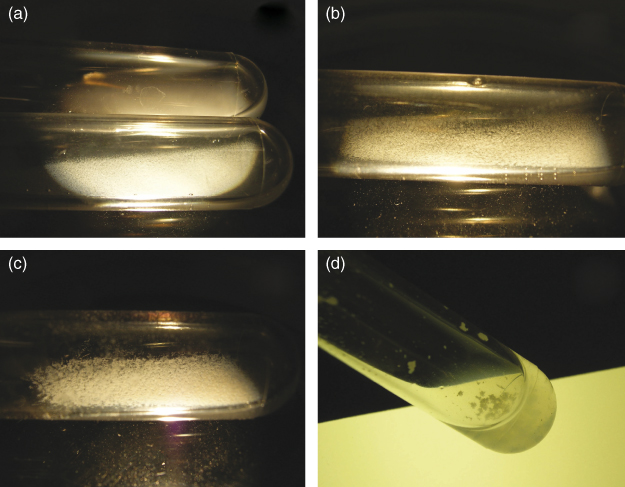
Figure 3.11 Coaggregation assay scoring. (a) Upper tube: “0” no change in turbidity and no evidence of coaggregates in the mixed suspensions; lower tube: “+1” turbid supernatant with finely dispersed coaggregates that did not precipitate immediately; (b) “+2” definite coaggregates easily seen but suspension remained turbid without immediate settling of coaggregates; (c) “+3” slightly turbid supernatant with formation of large precipitating coaggregates; (d) “+4” clear supernatant and large coaggregates that precipitated immediately.
(Reprinted from Journal of Endodontics, 32(10), Johnson EM, Flannagan SE, Sedgley CM, Coaggregation interactions between oral and endodontic Enterococcus faecalis and bacterial species isolated from persistent apical periodontitis, 946–950, © 2006, with permission from Elsevier.)
In contrast to synergistic relationships, interactions between different species and strains of the same species can be antagonistic. For example, some microorganisms produce antimicrobial proteins or peptides collectively termed bacteriocins that can be bacteriostatic or bactericidal to other members of the same species (narrow spectrum) or across genera (broad spectrum). Nonvirulent bacteriocin-producing strains have useful applications in the food industry and as probiotics (118). Bacteriocin production is not uncommon in enterococcal strains recovered from endodontic infections (48, 119). However, although the role of bacteriocins in RC biofilms is not known, it is feasible that by changing the composition of the microflora, the production of bacteriocins could modulate the infectious process. This was highlighted by the analysis of the strain E. faecalis MC4, originally recovered from an autogenously infected RC of a Macaca fascicularis monkey with apical periodontitis (120) and used as part of strain collections in seminal endodontic microbiology studies in monkeys (120–124). In those studies, E. faecalis MC4 survived in RCs for at least 2.5 years both as a single species and in combination with other species of an “eight strain collection,” and was shown in bacteriologic, radiographic, and histological studies to induce pulpal and apical pathogenesis (inducing the least immune effect when inoculated as a single species). Dahlen and colleagues (119) reported that E. faecalis MC4 inhibited other members of the eight strain collections to various degrees. More recent work has shown that the inhibitory activity by E. faecalis MC4 is associated with a Class IIa bacteriocin encoded by a pheromone-responsive, multiantibiotic resistance plasmid (125, 126).
In the oral environment, saliva flow impacts the development of biofilms on oral surfaces. However, the RC environment is neither subject to fluid flow nor necessarily fluid-filled. It has been hypothesized that during the initial phase of infection, exudate derived from the inflammatory lesion front may provide the fluid phase during the development of the RC biofilm (16). As such, in those cases where the biofilm is invading vital pulp tissue, nutrients would be available. In contrast, nutrient deprivation would be more likely to occur in RCs containing necrotic pulp tissue and in previously treated RCs. Many microorganisms manage to survive these challenging environments by prompting a starvation response whereby bacteria adjust their metabolic balance away from multiplication and toward the acquisition of energy for survival (127). Therefore, the adoption of a nongrowing state is an important mechanism for survival in the nutrient-deprived milieu of the RC.
As the biofilm progresses apically, it encounters the host’s complex and intricately coordinated immune system. Polymorphs attempt to wall off the bacterial biofilm from the remaining canal lumen (128), as can be seen in Figure 3.12 (90). However, in-vitro studies have confirmed that biofilms can eventually overwhelm the invading neutrophils (129) and reduce host defense responses as evidenced by a reduced cytokine release from macrophages exposed to E. faecalis in biofilm compared to planktonic conditions (130).
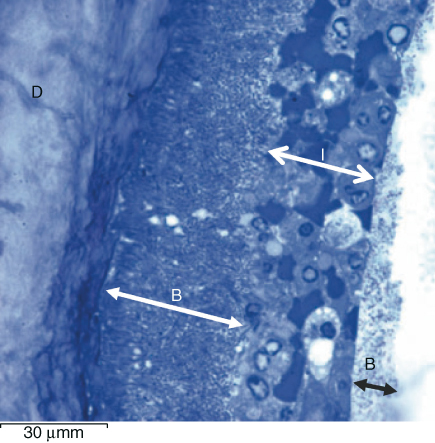
Figure 3.12 Light microscopic view of apical section showing bacterial biofilm (B) adherent to the canal surface dentin (D) and walled in by polymorphonuclear leukocytes and red blood cells (I) beyond which is a further biofilm (1 bar represents 30 µm).
(Richardson et al. (90). Reproduced with permission of John Wiley and Sons, Inc.)
Several studies have reported that there are a larger number of species (99) and more diversity (98) in acute compared to chronic infections (Figure 3.9). For example, black-pigmented gram-negative anaerobic bacteria such as P. endodontalis and P. gingivalis have been associated with symptomatic-infected RCs and endodontic abscesses (131–134). Molecular fingerprinting of 20 paired samples of RC and AAA exudates showed that, although the mean number of species and the diversity index did not differ, very few species were shared between RC and AAA samples, and microbial communities diverged between subjects (135) (Figure 3.13). Cluster analysis of the same clinical material showed that anaerobic species were distributed in small, complex, and distinctive communities in both RC infections and AAAs, indicating that several microbial profiles can be associated with the development of acute endodontic infections (103).
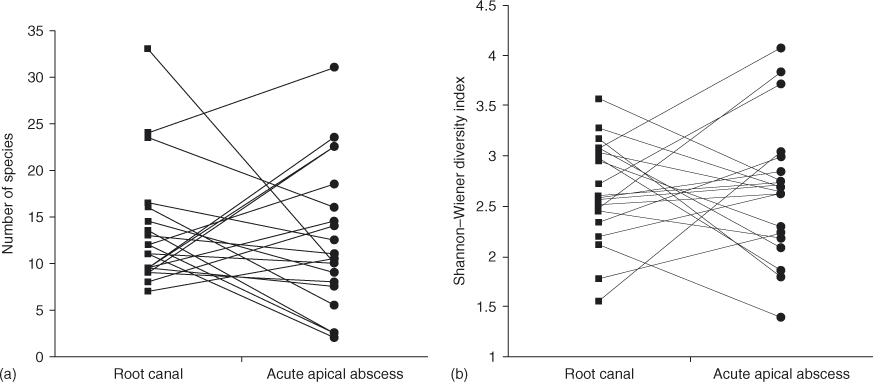
Figure 3.13 Microbial profiles associated with 20 paired samples of root canals (RC) and acute apical abscess (AAA) exudates. (a) Associations between RC and AAA samples in the number of species; (b) diversity index. Pairs of samples are connected by matching lines.
(Reprinted from Journal of Endodontics, 36(9), Montagner F, Gomes BP, Kumar PS. Molecular fingerprinting reveals the presence of unique communities associated with paired samples of root canals and acute apical abscesses, 1475–1479 © 2010, with permission from Elsevier.)
A classic study on infected RCs in monkeys showed that the microflora was more anaerobic in the apical compared to the coronal part of the canal (122). This is supported by a recent study that used pyrosequencing methods to compare the microbial ecology of the coronal and apical segments of infected RC systems of 23 extracted teeth with apical periodontitis (96) (Figure 3.14). The apical segment harbored more taxa overall and more fastidious obligate anaerobes, leading the authors to conclude that the apical part of the RC system drives the selection of a more diverse and more anaerobic community than the coronal part.
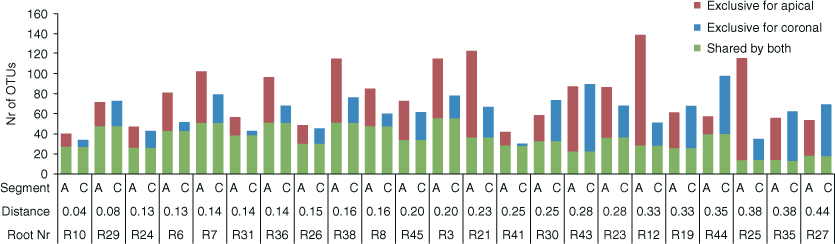
Figure 3.14 Prevalence of exclusive and shared operational taxonomic units (OTUs) (sequences that cluster within 97% similarity level) by the two root segments of 23 root pairs. Only those OTUs that contained at least 10 reads/sample were counted as present in the corresponding sample. The order of the samples/>
Stay updated, free dental videos. Join our Telegram channel

VIDEdental - Online dental courses


