Fig. 2.1
Classification of cancer biomarkers (Reproduced from Mishra and Verma [36] (http://creativecommons.org/licenses/by/3.0/))
Organ-/Tissue-Based Cancer Biomarkers
Oral Head and Neck Cancers
Of the numerous signature changes emerging as candidate biomarker for oral/head and neck cancers in recent years, mutations in the tumor suppressor p53 gene arguably stand out as the most studied to date. Protein of mutated p53 gene has been identified in saliva and in surgical margin analysis in head and neck squamous cell carcinoma (HNSCC) [47]. Recently, we reported the potential of MMP-9 and dentin sialophosphoprotein (DSPP) expressions at histologically negative resection margins of primary oral squamous cell carcinoma (OSCC) as predictive of primary site recurrence [48]. In an earlier report, DSPP expression in epithelial dysplasia of oral premalignant lesions indicated a fourfold increase in the likelihood of subsequent transition to invasive OSCC within 2 years compared to oral epithelial dysplasias lacking DSPP expression [49]. Loss of heterozygosity (LOH) and microsatellite instability at chromosome 3p, 9p, 17p, and 18q together with a hypermethylated promoter region of the p16 gene also are frequently cited as signatories to oral/head and neck cancers [36, 50]. There is as yet, however, no potential biomarker for oral/head and neck cancers of a caliber (prognostic/predictive value) comparable to the likes of ER/HER2 for breast cancer or c-KIT for gastrointestinal cancers. For this reason, it has been suggested that genomic profiling studies are needed to uncover head oral/head and neck cancer markers with prognostic/predictive values comparable to ER/HER2 or c-KIT for breast and GI cancers, respectively [36].
Lung Cancer
CEA is the most commonly used serum marker of lung cancers. Others are neuron-specific enolase (NSE), CA-125, cytokeratin fragment 21 (CYFR A 21–21), chromogranin A, retinol-binding protein (RBP), α(alpha) 1-antitrypsin, and α(alpha)1-antitrypsin [50]. Overexpression of K–ras, myc, EGFR, and Met oncogenes, or the inactivation of tumor suppressor genes, including p53 and Rb, has been cited as biomarkers for lung cancers [52–56]. Other reports alluding to the amplification of TTF–1, Pax9, and Nkx–8 at the DNA level as potential biomarkers of lung cancers have also been reported [36]. Within the last decade, it has been reported that hypermethylation of p16, RARB, and DAPK genes as detected in saliva and serum may predict the development of lung cancer [36, 57].
Uterine and Cervical Cancers
The role of high-risk HPV (HPV-16 and HPV-18) infection in the etiology and pathogenesis of cervical cancers has long been established. The E6 and E7 of the high-risk HPVs are the two oncoproteins whose cell cycle activity in the pathogenesis of cervical cancer has been extensively studied and fairly well understood and therefore pass muster test as uterine and cervical cancer markers [58]. Severely dysplastic uterine cervix lesions overexpress mini-chromosome maintenance (MCM) proteins, whereas overexpression of cycle protein 6 (CDC6) in malignant cervical cancer has been reported [59, 60]. The overexpression of p16INK4A has emerged as a favorite surrogate marker of preneoplastic and neoplastic uterine cervix lesions [61]. In addition to serving as a screening marker of early disease, the ribosomal protein S12 gene has been suggested as holding promise as a potential target in cancer gene therapy trials [62, 63].
Breast Cancers
A large number of breast cancer biomarkers have emerged within the last three decades and more continue to be held as promising suspects. At the moment, the American Society for Clinical Oncology (ASCO) has cataloged about eight protein-related tumor markers for breast cancer: CA 15–13, CA 27–29, carcinoembryonic antigen, estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2), urokinase plasminogen activator (uPA), and plasminogen activator inhibitor (PAI)-1 [36, 64]. These subserve different aspects of breast cancer activities: monitoring (CA 15–13, CA 27–29 and CEA), treatment planning (ER, PR, and HER2), and recurrence/risk prediction (uPA and PAI-1) [64]. p53, cathepsin D, cyclin E, and kallikrein 14 have now been added to the list as protein-based potential biomarkers of breast cancer [36, 64, 65].
Messenger RNA (mRNA) expression-based platforms such as MapQuant Dx™ Genomic Grade (based on the expression of about 100 genes for breast cancer detection) and the RT-PCR-based assay system, BCtect™ with several genes for early detection, are now available [66]. There are also reports suggesting that a number of microRNAs (miRNA), including mir-125b, mir-145, mir-21, and mir-155, known to be dysregulated in breast cancers be considered among the list of candidate biomarkers for breast cancers [66, 67]. Indeed, reviews of the gene-expression signatures, namely, mRNA transcript expression patterns, have formed the bases for the classification of treatment modalities for breast cancer patients [36]. Hypermethylation of BRCA1, p16, and 14–3–3 σ(sigma) presents in about 95 % of sporadic breast cancers, and genes coding for cyclin D2, and RAR–β(beta) represent epigenetic biomarkers [68]. Other candidate and proposed biomarkers for breast cancer reported to date include: serum HER2; cytokeratins 8, 18, and 19; kallikrein; osteopontin; mutant p53; and crypto1. Effort to identify additional breast cancer biomarkers is being enhanced by the availability of nanotechnology [69].
Liver Cancer
To date, the frontrunner biomarkers for liver cancer (hepatocellular carcinoma) are alpha-fetoprotein (AFP) and its derivative, AFLP (Lens culinaris), and des-carboxy prothrombin (DCP) [70–72]. Proposed markers include plasma levels of transforming growth factor beta 1 (TGF-β1), I-6/10, IGF, and gamma-glutamyl transferase (GGT) enzymes [73, 74], while less established markers include levels of glypican-3 (GPC3) and golgi protein 73 (GP73) [36, 73, 74].
Prostate Cancer
In spite of the popularity and frontline status that PSA has enjoyed over many decades as a biomarker for prostate cancer, its validity has come under close scrutiny in recent years. This is because there is now increasing evidence, including data from the American College of Surgeons National Cancer Data Base (NCDB), showing that not all patients with previously acknowledged PSA levels associated with prostate cancer have cancer [36]. Furthermore, increasing knowledge of the interplay between ratios of the different PSA subtypes (fPSA, tPSA) in predicting such factors as the aggressiveness of prostate cancer has contributed the eroding the validity of PSA as a cancer biomarker [75]. This turn of events has served to refocus effort and attention to the investigation of other potential candidate biomarkers for prostate [36]. Candidate markers now touted include the overexpression of human kallikrein-related peptidase 2 (hK2), early prostate cancer antigen (EPCA), α(alpha)-methylacyl-coA racemase (AMACR), insulin-like growth factors and binding proteins (IGFBP-2 and IGFBP-3), and TGF-β(beta)1. Others are elevated circulating levels of interleukin-6 (IL-6) and its receptors, urokinase plasminogen activator (uPA) and receptor (uPAR), enhancer of zeste homolog 2 (EZH2), and prostate-specific membrane antigen (PSMA) [76–83].
Infectious Agent Cancer Markers
Viral Markers
The roles of infectious agents, notably viruses, in the etiology and/or pathogenesis of specific human cancers are fairly well established. Spanning over nearly half a century now, the link between Epstein-Barr virus (EBV) and a number of human malignancies, including nasopharyngeal carcinoma and Burkitt’s lymphoma, is well documented [84, 85]. The established causative relationship between oncogenic HPVs (HPV-16, HPV-18) and uterine cervix cancer has already been discussed above, and similar potential etiologic relationship with a subset of oral/oropharyngeal cancers remains an area of intense investigation. Although HPV-16 (and to a lesser frequency HPV-18) is now easily demonstrable in a subset of oral cancers, proof of etiologic partnership between high-risk HPVs and oral cancer is less than concrete [86]. Hepatitis B virus (HBV) and hepatitis C virus (HCV) most probably have causal relationships with hepatocellular carcinoma [87]. Within the last three decades, the human herpes virus-8/Kaposi’s sarcoma herpes virus (HHV-8/KSHV) has been firmly associated with Kaposi’s sarcoma (KS), other sarcomas, and certain lymphoma [64]. The RNA virus, human T-cell lymphotropic virus type 1 (HTLV-1), is regarded as an etiological factor for certain types of leukemia [88].
Bacterial Markers
Helicobacter pylori is probably the only well-characterized bacterial pathogen associated with a specific human cancer. Although essentially an infectious agent whose primary pathogenic activity manifests by way of inflammation and ulceration of the upper gastrointestinal tract, including the stomach (gastrum), the last two decades have seen the apparent acceptance of H. pylori as a solid biomarker for gastric cancer [89, 90]. However, it is estimated that over 50 % of the world’s population harbor H. pylori in their upper gastrointestinal tract with infection being more prevalent (80 %) in developing countries [36]. This certainly casts some shadow regarding the timeline significance of H. pylori detection in patients as well as its clinical utility and validity as a biomarker for gastric cancer. Either DNA polymorphisms or antibody-based technologies are used to detect H. pylori in patients [36]. The significance of H. pylori is perhaps more as a surrogate risk factor and therefore a surveillance parameter for monitoring potential disease progression, than it is a biomarker for gastric cancer.
Blood Versus Saliva as Diagnostic Medium
Serum and urine are still regarded as the gold standard body fluid for clinical investigations involving proteomics and the biochemical assays of other body fluid constituents. Even with advances in clinical proteomics providing the opportunity to identify disease biomarkers in biological fluids at reduced costs for early diagnosis of disease, most of the focus remains on the proteomics of serum or plasma. Predictably, the advent and rapid advances of the past two decades in the field of salivary diagnostics and the rising profile of saliva as a diagnostic medium are beginning to generate debates on a couple of fronts. The first questions whether saliva will ever be ready for “primetime” as a diagnostic body fluid; the second, and much more fundamental, is whether, aside from intellectual curiosity, there is a real need for a second (or third) diagnostic body fluid or the need to supplant serum/plasma at this time.
The Case for Saliva (Over Serum)
The attention toward other body fluids, such as saliva and urine, as attractive alternatives of potential sources of disease biomarkers, and as diagnostic fluid, is because of the noninvasive processes associated with these alternatives. The obvious attraction of saliva as a clinical tool in diagnostics is its availability (in the absence of xerostomia/dry mouth due to chronic disease state) as well as the relative ease of collection and storage compared to serum and urine [91, 92]. The process of saliva collection is relatively straightforward and convenient, particularly in multiple samples situation [91, 92]. In stark contrast to the “invasive” process of collecting blood/serum from patients, the noninvasive process of saliva collection is near absolute. For example, patients who are phobic to needle pricks that invariably involved for blood draws will readily accede to the friendlier procedures for saliva collection, which is largely devoid of threatening maneuvers and also compatible with a stress-free appointment. Furthermore, the fact that, unlike blood, saliva lacks clotting ability simplifies the preservation steps and makes it easier to handle.
It is estimated that 20 % more Americans visit the dentists regularly than they do their physicians in any given time [12, 93]. This positions the dentist as an enhanced gateway opportunity for early detection systemic diseases, including life-threatening conditions [93]. Also the results of a survey by Greenberg and colleagues of 1,900 practicing dentists in the United States to determine their willingness to obtain saliva samples from patients and send same to a laboratory for diagnostic evaluation showed an 87 % “yes” response from respondents [93]. This result provides some evidence of the receptiveness of the dental profession to saliva-based diagnostic and risk-assessment technologies [12, 93]. From the perspective of dentistry as a profession, a full integration of salivary diagnostics into the practice of dentistry will enhance its profile and significance in the overall scheme of primary health care [12].
Given the present state of salivary diagnostics technology, it would appear that for routine clinical utility, the technology is currently expensive compared to the established serum technology. However, it is anticipated that as salivary diagnostics achieve a validity threshold, and subsequently becomes commonplace, its cost-effectiveness will become favorable. Recent development of room temperature analyte stabilization methods, nucleic acid pre-amplification techniques, and direct saliva transcriptomic analysis for accurate detection and quantification of saliva transcripts have already proven to be cost-effective [94]. Indeed, the lack of commercial success of serum-based markers have been deemed as presenting an opportunity for salivary diagnostics to thrive commercially, more so because several salivary biomarkers for oral and systemic diseases are poised for translational and clinical applications, and the market may well be ready for a “a new molecular-diagnostic product” [94]. For remote and impoverished regions and communities, salivary diagnostics offers affordable, yet highly accurate, alternative to the more expensive serum technology [94].
An anticipated utility, and therefore advantage, of salivary diagnostics technology is in the fight against bioterrorism [95]. Under these scenarios, a robust, fast, portable, and user-friendly technology with the capacity to screen for multiple agents simultaneously will offer a distinct advantage [95]. Saliva and other oral fluids collected for this purpose following defined forensic guidelines, including the necessary observation, will be less susceptible to adulteration [95]. In summary, the advantages of saliva as a clinical tool over serum are: the noninvasive procedures involved with obtaining sample, a smaller sample (aliquot) requirement, good patient psychology and cooperation, relative cost-effectiveness at the full turn of the requisite technology, ease of storage and transportation, greater sensitivity, and correlation of result analysis with that of serum.
The Case for Serum (Over Saliva)
While saliva diagnostics for detection of oral diseases (caries, periodontal disease, oral cancer, salivary gland disorders) makes natural sense to many, its reliability and utility in the diagnosis of systemic diseases remote from the oral cavity have been deemed as speculative by just as many [12, 95]. Among many others, a commonly adduced reason for the skepticism, and therefore the reluctant acceptance as an emerging field, is the absence of a clear mechanism providing the nexus between the expression of certain substances in salivary and a disease state at a remote organ or body site [12]. To address the mechanistic question, animal models are proposed with several investigators already exploring this using rodent tumor models [12, 95]. For example, one working hypothesis investigated via the animal models is that exosomes, which are microvesicular structures (30–100 nm in size) shed by tumors, transport tumor-specific contents to distant sites, including the salivary glands, accounting for their remote presence as “disease-discriminatory markers in saliva” [96].
Saliva as a diagnostic fluid is yet to achieve wide acceptance despite the gains and associated generous publicity of the last two decades. Although the same period of time has witnessed the approval of salivary diagnostics-related products by the FDA, such as the saliva HIV antibody detection kits and a number of saliva-based kits for detection of hormones, clinical test application remains quite minimal [3, 12]. It has been suggested recently to be partly attributable to the portrayal of saliva in a negative light in some quarters and culture where social, cultural, and psychological stigmas are associated with saliva [3]. Therefore, it is conceivable that in such cultures, the collection of saliva in even in a clinical setting may be equated to “spitting in public” [95].
Relative to the current state of technological and infrastructural support for salivary diagnostics, the infrastructure of clinical laboratories and blood-serum detection assays are already well established to argue against any need for an equivalent whose competitiveness, viability, and validity are yet to be fully assessed [3]. It is further contented that, unlike the well-established serum technologies that already offer reliable tests for early detection and diagnosis of diseases such as cancer, the current stage of development of salivary diagnostic technology is still years from attaining the same level of reliability [3, 12]. In summary, further in-depth understanding of the nexus between biomarker of systemic diseases and the salivary gland and the validation of current salivary diagnostics technology via multiplex assay are hurdles on the path of universal acceptance of saliva as a diagnostic fluid. This fact also suggests that, because of the need to substantiate the scientific rationale of salivary biomarkers for systemic diseases, that the pace of acceptance of saliva as a diagnostic medium by physicians may be slower than that of dentists for oral diseases [3].
Legal Issues Related to Salivary Diagnostics
There does not appear to be available as yet an established legal framework specifically focused on salivary diagnostics, even though the use of saliva (and oral fluids in general) for forensic and drugs of abuse investigations is fairly well established. In this section, an attempt is made to summarize and speculate on potential areas of legal ramifications that may arise from the use of saliva for clinical diagnosis of human diseases. Some of the anticipated bases for challenges include: the mode of saliva sample collection, accuracy/reliability/validity questions regarding saliva test outcomes, challenges to suitability of test site, specimen integrity (including chain of custody), and interpretation of test results.
Scientific Evidence in the Legal System
A primary hurdle that scientific evidence in the US legal system must cross is satisfying the legal requirements for “admissibility.” Essentially, admissibility speaks to the full “evidentiary value” of any scientific evidence via a process of determining whether or not the evidence is reliable enough to be entertained by the court and subsequently admitted as part of the court record for the issue at stake. On admission, the “weight” to be ascribed to the evidence in the context of the matter is assessed. The standards of evidentiary proof are hardly consistent and will depend on whether the proceeding is adversarial/non-adversarial. In adversarial proceedings, for example, evidentiary proof ranges from “beyond a reasonable doubt” in criminal cases to “preponderance of the evidence” in civil matters or probation hearings. Lower standards of proof, such as “probable cause” and “reasonable suspicion,” are occasionally entertained. Thus, the scientific certainty of test results is subject to different levels of scrutiny depending upon the nature of legal proceedings as indicated above. Figure 2.2 summarizes estimates of percent accuracies used for various legal decision-making [4].
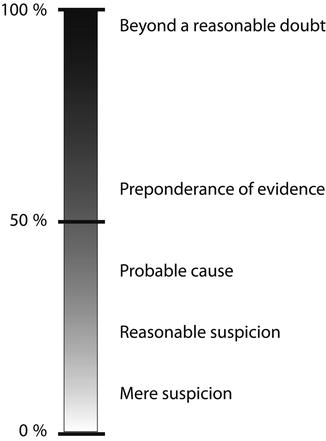

Fig. 2.2
Hierarchy of legal decision-making (Modified from Kadehjian [4] with permission from Elsevier)
Admissibility of scientific evidence in the US federal courts is governed by the 1975 Federal Rules of Evidence [4, 97]. In 1993, the US Supreme Court used the opportunity of the Daubert v. Merrell Dow Pharmaceuticals Inc., 509 U.S. 579 [98] (and other cases that followed shortly) to elaborate and provide guidance on the criteria for determining the admissibility of scientific evidence [97]. In assessing the admissibility of scientific evidence, the US Supreme Court, via the Daubert case, enumerated the following factors that could be considered by a judge in answering admissibility questions related to scientific evidence: testing, peer review or publication, known or potential rate of error, standards controlling operation, and general acceptance [98]. Currently, about two-thirds of the US states have adopted the Federal Rules of Evidence as it pertains to scientific evidence [4]. Although the use of saliva and other oral fluid tests for drugs of abuse appears to routinely meet the requirements, it is unclear how well salivary diagnostics will fair, particularly with respect to the factors enumerated in the Daubert case. At least, it is reasonable to anticipate the courts to accord deference comparable with that in saliva tests in drug abuse cases to legal challenges of equivalent aspects of salivary diagnostics testing.
Potential Legal Challenges to Salivary Diagnostics
The acceptability of saliva as a specimen for analysis is borne out by both history and a record of acceptability [4]. Saliva tests are now commonplace in forensic investigations when samples may be obtained from drinking glasses, cigarette butts, envelopes, and other sources to detect blood-group substances, salivary genetic proteins, and many other substances [3, 4]. However, as is characteristics of formative stage projects, part of the uncharted waters of salivary diagnostic are potential legal landmines that may accompany its increasing relevance. It is prudent to anticipate that with stronger footholds of salivary diagnostics comes the likelihood of legal challenges, particularly if sanctions are a possible outcome of legal resolution. Although it is impossible to see all the facets of potential legal questions to be generated by salivary diagnostics from our current vantage, likely areas include challenges to: insurance claims related to positive/negative saliva tests, methods of saliva sample collection, accuracy and reliability of laboratory test results, and interpretation of test results.
Challenges to insurance claims (post-treatment or preapproval for therapy) based, for example, on positive or negative results of salivary diagnostic tests are foreseeable. Insurance companies may deem saliva test results and diagnosis unacceptable for reimbursement purposes or to approve therapy and may insist on verifications with a “gold standard”—presumably serum. In litigation, questions also may arise regarding specific methods of collection. A method of collection must be consistent with preservation of the integrity of the specimen before tests. As an illustration, if the use of citrate to stimulate saliva flow could be proven to, even remotely, interfere with assayed substances, results of such tests may prove unreliable [4, 99]. The FDA approval/validation of both methods of saliva collection and laboratory procedures for assays is likely to be a requirement for insurance reimbursement. Even when the saliva tests are deemed accurate, challenges may be focused on interpretation of the results [4]. For example, while the presence of a particular substance in serum may be “diagnostic” of the disease in question, its presence in saliva (outside the serum evidence) may not necessarily meet the required diagnostic threshold for the same disease.
Stay updated, free dental videos. Join our Telegram channel

VIDEdental - Online dental courses


