Pooled sample studies are working toward identifying a global oral microbiome profile for healthy individuals. One study found 72 % of the genus level or above, which accounted for 99.8 % of all the organisms present in the mouth, was the same among unrelated healthy volunteers [
16]. Another group found that 15 genera were found to overlap between 10 unrelated healthy individuals [
40]. Of these genera, 13 were found to be in common between both studies:
Streptococcus,
Corynebacterium,
Neisseria,
Rothia,
Veillonella,
Actinomyces,
Granulicatella,
Fusobacterium,
Haemophilus,
Prevotella,
Campylobacter,
Capnocytophaga, and
Cardiobacterium [
16,
40]. This overlap creates an outline of the typical organisms and their relative amounts present within healthy individuals, defining the so-called oral “core” microbiome.
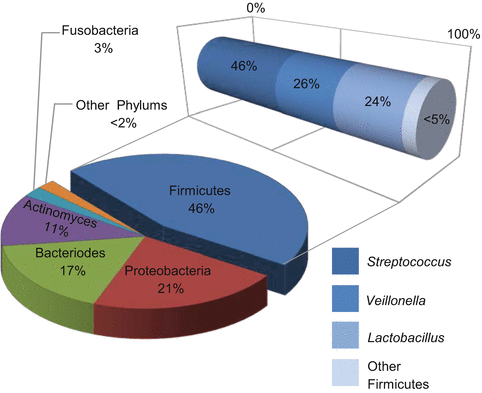
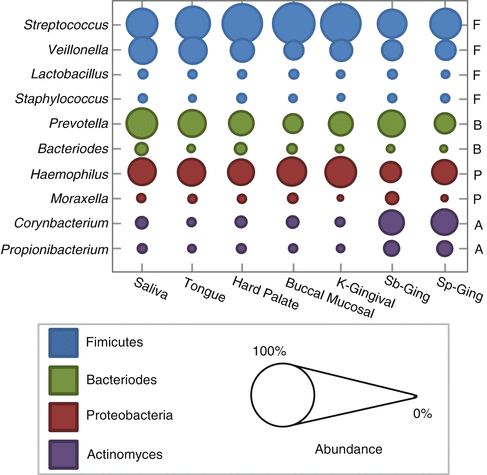
Thorough evaluation of oral micro-niches has recently become possible thanks to the culture-independent methods of high-throughput sequencing. Multiple studies have been published outlining the normal microorganisms found in various locations in the mouth. Analysis in one study revealed that at each oral site tested, there was an average of 200–300 different “species-level” phylotypes [16]. Interestingly, comparison between these unrelated healthy volunteers showed a significant overlap in the typical organisms present in each location. When samples were compared across various sampling sites in the mouth, saliva showed the closest relationship to the tongue first, then palate, mucosa and gingiva, and finally plaque [3, 15, 16, 32, 38, 47] (Fig. 5.2). Firmicutes were the dominant species in saliva followed by Bacteroidetes, Proteobacteria, and Actinobacteria [16, 32, 34, 36]. Together, these four phyla made up >96 % of the total bacterial composition. Salivary samples were also found to be relatively stable in composition over time among healthy individuals [2, 36, 48, 49]. Samples taken at three separate time points over a period of 29 days showed consistent results in their overall salivary microbiome profile [36]. Overall, these studies revealed that each oral location tested resulted in a relatively unique profile or relative number of microorganisms and these healthy communities are stably maintained over time.
This persistent maintenance of the “normal” flora appears to be a key feature not only in the oral cavity but also in the gut, skin, and vaginal locations [
2–
4,
11,
15,
16,
31,
48,
50]. Strikingly, the oral environment was found to have the most consistent “core” microbiome maintained between unrelated people in comparison to other microbial sites such as the gut [
2,
36,
40]. The other microbiome sites in the body showed a diverse population of bacteria, but there was considerable variation between unrelated individuals [
2,
40]. Defining this “core” microbiome is one of the primary steps that allow for future comparison studies to define profiles that shift away minimally or aggressively from the normal profile.
Shaping the Normal Oral Microbiome
Oral bacteria have preferences for specific sites in the mouth including the teeth, tongue, hard and soft pallet, buccal mucosa, tonsils, gingival tissue, and saliva. Several factors influence the formation and maintenance of the oral ecosystem including the host (host tissues, fluids and signals, diet, genetics), the local environment (pH, temperature, oxygen, amount of nutrients), and the microorganisms themselves (adherence, coaggregation, inter-/intraspecies interactions, virulence mechanisms). The particular community of microorganisms is normally maintained in a state of symbiosis with the host. Yet this diversity can be shifted with changes in the local host condition along with the influence from beneficial and/or antagonistic microbial interactions. In fact, many oral pathogens are opportunist. These pathogens only show virulence when presented with a susceptible host or when a normal host undergoes a number of changes within their oral physiology. Multiple studies have shown that microbial species show distinct properties when part of a multispecies community versus growing in isolation [
51–
59]. Specifically, work by Foster and Kolenbrander [
54] showed that the adherence of certain oral species to an artificial tooth surface was dependent on specific microbial binding partners. These multispecies communities generally respond to changes as a group. The microbial population is continuously undergoing a process of turnover. New microorganisms colonize and persist at the expense of others and based on complex interactions will then influence subsequent population compositions [
57].
The foundation of these oral communities starts with the deposit of the salivary pellicle on surfaces. The pellicle is primarily composed of glycoproteins derived from saliva that are recognized by bacterial adhesions, allowing selective binding of mainly Gram-positive cocci to the pellicle surface. In addition to providing a binding surface for bacteria, saliva also provides nutrients for bacterial growth from both endogenous (glycoproteins) and exogenous (carbohydrates and peptides) sources. The saliva also shapes the composition by introducing antimicrobial elements and clearance of bacterial species (see later). Bacteria that have attached to the pellicle generate bacterial products as they grow and multiply that influences the subsequent colonizers. A number of organisms are dependent on coaggregation in order to form part of the microbial community [
57]. Once the initial microbial population is established, there is a shift observed in composition as more organisms attach and also based on growth rate differences. The majority of these bacterial communities must be constantly rebuilt in response to the mouth’s natural cleansing activities such as saliva production, abrasion, and swallowing.
The Role of Saliva in Forming the Oral Microbial Environment
Saliva is a complex mixture of salivary gland secretions, gingival crevicular fluid, microorganisms, microbial by-products, epithelial cells, and other chemical components. Saliva is secreted by three primary glands, the sublingual, the parotid, and the submandibular, as well as by many minor glands [
60]. The amount and type of components present in saliva are known to affect many aspects of oral health and influence bacterial growth [
19,
54,
57,
61–
66]. The bicarbonates, phosphates, and urea within saliva act to modulate the pH and buffer the oral cavity. Salivary proteins contribute to oral microbial metabolism, aggregation, and attachment as well as bacterial cleansing. Immunoglobulins, enzymes, and proteins in saliva manage bacterial growth through antimicrobial action. Finally, saliva is primarily composed of water and therefore provides the necessary moisture for bacterial survival.
The normal pH of saliva is slightly acidic from 6 to 7; however, the range varies based on salivary flow, with values near 7.8 during high flow, and the pH can approach 5.3 at low flows [
60]. Bacteria are able to survive in a wide range of pH conditions, but the healthy microbiome composition is primarily supported by a neutral pH range [
19]. Circulating bicarbonate and phosphate systems work together to buffer saliva and maintain pH levels [
60]. Bacteria also help to buffer saliva by breaking down urea to ammonia and CO
2, resulting in an increase in pH [
67,
68]. The buffering action of saliva is greatly dependent on high flow rate, allowing regulation of the pH in regions such as the oral plaque environment on and around the teeth [
69]. The amount of saliva produced in the mouth displays regional variation, with different areas of the mouth producing various volumes. This variation in saliva volume has been directly linked to the regional clearance rate of acid produced by oral bacteria, generating pH micro-niches within the oral cavity [
60]. This variation explains some of the regional differences in bacterial composition seen in the oral cavity (Fig.
5.2). A shift toward acidic pH is known to support the growth of acid-producing and acid-tolerant organisms while killing off acid-susceptible bacteria.
There is a large variability in individual flow rates of saliva [
70]. Salivary flow is commonly triggered by mechanical chewing and the taste or smell of food. Other factors that influence salivary flow rates are pain, certain medications, and various local and systemic diseases [
71–
76]. The salivary glands are linked to the sympathetic and parasympathetic nervous system [
77]. There are also a number of neurotransmitters and hormones that can influence the rate of salivary flow. Insufficient salivary flow has been clearly associated with a significantly increased risk of oral disease [
64,
72,
74,
78,
79] coinciding with a shift in the oral flora [
55,
61,
72,
80,
81] (see later). Nutritional changes and deficiencies have also been shown to affect salivary function [
19,
82–
84] (see later). Salivary flow rates correlate with clearance of fermentable carbohydrates and buffering around high-dental-plaque zones.
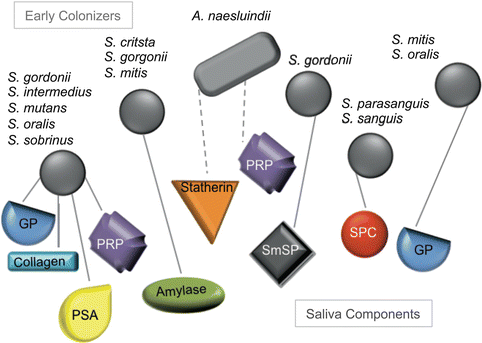
Colonization of oral surfaces by bacteria depends on binding to the salivary pellicle (Fig. 5.3). The acquired salivary pellicle is a proteinaceous layer that covers all exposed surfaces in the oral cavity. This complex layer is composed of approximately 90–130 different proteins derived from saliva and crevicular fluid serum [60]. These proteins have been grouped based on various roles including binding calcium and/or phosphate, interaction with other proteins, antimicrobial activity, inflammatory response, immune defense, lubrication, and/or buffering and remineralization. Pellicle formation depends in part on the presence of intact mucins and proline-rich proteins (PRPs). Mucins are glycoproteins mainly secreted by sublingual, submandibular, and palatal glands [87–89]. PRPs constitute the majority of the human parotid saliva [90]. Mucins and PRPs help initiate colonization by healthy oral flora and are incorporated into the bacterial biofilm structure [87, 88, 91, 92]. Healthy bacteria and the mucin pellicle component provide a protective layer that shields against acids and excessive wear of oral surfaces [88, 93]. If excess colonization takes place, mucins are able to aggregate oral bacteria and clear them from oral surface [93]. Mucins are also critical in hydration and lubrication in the oral cavity and thought to play a role in binding toxins [93–95].
The antimicrobial activity of saliva is primarily maintained by production of immunoglobulins and enzymes. Salivary glands secrete immunologic agents to protect the teeth and mucosal surfaces. IgA, IgG, and IgM are common immunological salivary components. Secretory IgA is the dominant immunoglobulin on all mucosal surfaces, and it works to neutralize viruses and to aggregate bacteria and functions as an antibody to bacterial antigens [
96,
97]. Mucin had been shown to work with secretory IgA and bind bacterial pathogens with greater affinity than either molecule working alone [
98]. IgM is thought to work in a similar way to IgA but is more susceptible to proteolytic degradation [
99]. IgG is primarily added to saliva via crevicular fluid and has been found to inhibit colonization of the oral pathogen
Streptococcus mutans [
100]. Saliva-based enzymes (lactoferrin, lysozyme, peroxidase, amylase) protect the teeth from microbial insults [
19,
60]. Nutritional immunity is enacted when lactoferrin binds salivary ferric iron, a necessary nutrient for some oral bacterial species [
101,
102]. Lysozyme destroys bacterial cell walls and also promotes clearance through bacterial aggregation [
19,
103]. Peroxidase catalyzes the production of bactericidal by-products such as thiocyanate [
104]. Amylase is a well-known digestive enzyme secreted by the parotid gland [
105]. It functions to modulate the adhesion of certain oral species (e.g.,
Streptococcus gordonii and
Streptococcus mitis) while forming part of the salivary pellicle and as a component in free saliva [
19]. Finally, amylase has been specifically found to inhibit the growth of bacterial pathogens
Legionella pneumophila,
Neisseria gonorrhoeae, and
Neisseria meningitidis [
106–
109].
Different salivary glands produce saliva with various levels of key salivary components [
87,
110]. Differences are seen in the growth of oral bacteria on saliva from different glands [
111]. This alone demonstrates that variations in saliva components are able to influence the microbiome of a healthy adult. At the outset, studies have looked at the direct effect on saliva composition and flow rates based on medication, radiation treatment, diet, or disease/trauma directly linked to the salivary gland functionality [
73,
76,
79,
101,
112,
113]. A classic example is seen with the autoimmune disease Sjögren’s syndrome. Patients with Sjögren’s have chronic inflammation that affects their salivary glands leading to reduced salivary flow [
76,
113]. Radiation has also been shown to decrease not only the salivary flow rate but also buffering capacity and pH [
79,
114]. Together, altering the buffering capacity, salivary flow, and/or protein levels can all lead to major changes in the oral environment.
The Dynamic Oral Microbiome: Shifting Bacterial Compositions in Response to Change
Changes in salivary flow can greatly influence the microbial composition present in the oral cavity [
54,
55,
60,
72,
80,
114]; pH buffering, bacterial nutrient access, and components of the host response all shape the oral environment [
19,
60]. While bacterial populations are able to remain relatively stable throughout the day with multiple shifts in saliva production levels and access to fermentable substrates, prolonged difference in the salivary composition or flow rate can result in a dramatic shift in the oral population. Bacterial mechanisms to tolerate external stress have been well documented [
172,
175–
177]. Cell-cell communication and biofilm formation oftentimes allow these microbial communities to quickly adapt and survive short-term extreme changes in the environment (e.g., lack or food, host antimicrobial factors). Yet, these complex communities inevitably demonstrate changes in species composition after persistent changes occur in the host environment. On the microbial scale, even minute changes that are not readily perceivable by the host, if consistent, will impact the microbial composition. These shifts in host-microbial homeostasis demonstrate that, instead of considering our microbiome as something peripheral, we need to recognize our flora as an integral part of our body systems.
Next, we will discuss how the oral microbiome shifts away from the oral core microbiome in response to oral-specific diseases and systemic diseases/conditions.
Diseases of the Oral Cavity
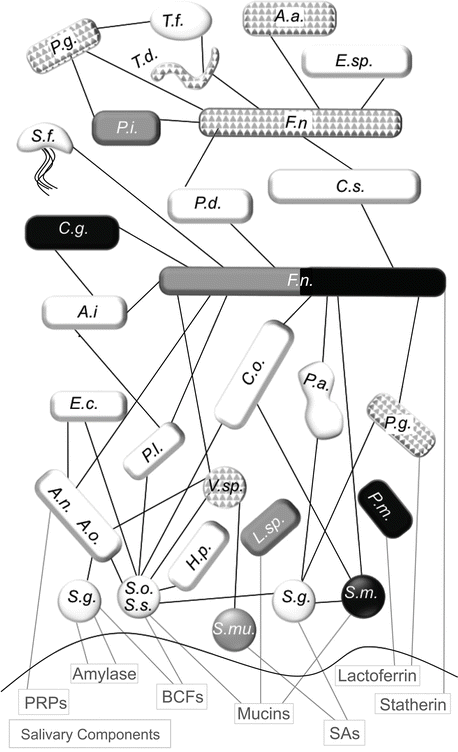
Some of the most well-documented relationships between health, disease, and the oral flora can be observed with the common dental ailments of tooth decay (caries) and periodontitis, including gingivitis. These diseases represent two of the most common chronic diseases seen worldwide [178–180]. Several decades of research by dental scientists have clearly illustrated that dental caries and periodontitis are caused by ecological changes in the homeostatic balance between host and bacterial flora (Table 5.2). While initial work assumed that the presence or absence of a single species would be linked to cause and effect for these diseases, it was quickly discovered that these diseases develop along a more complex story line. These oral diseases involve regulation by many sources such as multispecies bacterial connections, host-bacterial interactions, and behavioral or environmental shifts. This body of work begins to demonstrate that changes in the microbial balance can both lead to and result from active oral disease profiles in humans. Oral biofilms have been studied extensively in oral diseases. There is a general progression seen in the buildup of the oral composition as early/primary colonizers attach to the salivary pellicle, additional bacteria bind to the primary colonizers, and finally late colonizers join the group only after a complex community is present. These later colonizers include a number of key oral disease-linked bacteria.
Table 5.2
Bacteria connected to oral health conditions
|
Bacteria
|
Actinomyces spp.
|
Aggregatibacter actinomycetemcomitans
|
Actinomyces spp.
|
|
Lactobacillus spp.
|
Campylobacter rectus
|
Capnocytophaga sputigena
|
|
Propionibacterium acidifaciens
|
Fusobacterium nucleatum
|
Cardiobacterium hominis
|
|
Streptococcus mutans/sobrinus
|
Porphyromonas gingivalis
|
Haemophilus parainfluenzae
|
|
Veillonella spp.
|
Prevotella intermedia/nigrescens
|
Rothia dentocariosa/mucilaginosa
|
|
–
|
Treponema denticola/forsythensis
|
Streptococcus sanguinis
|
|
References
|
|
|
|
Caries
Dental caries lesions are localized demineralization of tooth enamel as a result of acid production by dental plaque biofilms. Caries progression has been well documented from superficial white spot lesion to deep-dentin cavitations [
56]. This decay is due to chronic acid production from bacterial fermentation of carbohydrates in the vicinity of the tooth enamel [
56]. Previous work looking for the causative agent of dental caries showed that mutans streptococci (
Streptococcus mutans and
Streptococcus sobrinus) appeared to be the primary acid producers associated with enamel caries [
56,
134–
136,
171,
181]. These bacteria were consistently shown to create the extreme acidic environment necessary for caries conditions, a pH <5.5 [
182–
184]. More recent research has shown that
Lactobacillus spp. are also prevalent at active caries site and appear to create the necessary cariogenic conditions in the absence of the
S. mutans or
S. sobrinus [
137,
138,
185]. These species are all aciduric, meaning that they thrive in low pH environments, while other members of the oral flora do not survive long exposures to acid conditions. The loss of competing species during low pH events creates a feedback loop that ultimately yields more acid production, due to the enrichment of aciduric species in the population, leading to the progressive destruction of enamel and dentin, if left untreated. Thus, caries patients display an increasing accumulation of these cariogenic bacteria. Modern techniques using sequencing technology have allowed for further characterization of not just a few select species but analysis of the entire microbiome at the disease sites. More detailed microbiome sequencing reveals an apparent distinct overall microbial shift as the tooth deterioration progresses from mild white spot lesion to caries dentin involvement and finally to severe disease with deep-dentin lesions [
21,
46]. The attachment of specific oral flora to the salivary pellicle was directly tied to the particular binding partners that were present or absent during multispecies biofilm formation [
51,
116]. Organisms that would normally be able to bind to the pellicle may be outcompeted for binding sites by other common oral species, while others are completely dependent on “helper” bacteria to mediate any active binding to the pellicle surface. These multispecies interactions appear to significantly influence the makeup of the biofilm composition as well as the circulating bacteria in saliva. These interactions must be constantly reestablished following removal from common oral practices such as swallowing, mechanical abrasion from mastication, and toothbrushing.
Lapses in oral hygiene and increased access to fermentable carbohydrates correlate with increased acid production from certain oral species leading to a localized drop in pH at sites with high levels of dental plaque [
83,
186,
187]. This drop in pH then starts an environmental feedback cycle shifting some of the oral flora in the mouth. The decrease in pH starts to kill off susceptible oral bacterial species and selects for acid-tolerant organisms while simultaneously dissolving mineral from the tooth enamel. As the caries disease progresses, the environment has been shown to enrich growth of acid-producing and acid-tolerant bacteria including
S. mutans,
S. sobrinus,
Propionibacterium acidifaciens,
Lactobacillus spp.,
Veillonella spp., and
Actinomyces spp. [
138,
188]. Allowed to persist, this localized loss of mineral from of tooth enamel can lead to significant structural deterioration and pain.
While these acid-tolerant species increase in numbers, it should be emphasized that the remaining oral species do not completely disappear, but instead are less prevalent at the disease site. It was mentioned earlier that the oral environment normally undergoes a number of environmental shifts throughout the day; nonetheless, the overall composition of the oral flora remains constant in healthy individuals [
177,
189]. Thus, bacterial and host biomarker profiles can then be used to infer the risk of developing dental disease as well as monitor dental treatment effectiveness. For example, caries risk has been assessed by the amount of lactobacilli, mutans streptococci, and
Actinomyces spp. in the patient’s saliva [
46,
135,
137,
139,
140,
185]. Young children with high levels of
S. mutans and
S. sobrinus were more than five times as likely to develop dental caries than individuals with lower levels [
135,
136,
141,
190]. Sweden has used salivary levels of lactobacilli (>100,000/ml saliva) and mutans streptococci (>1 million/ml saliva) to determine caries risk for more than 30 years [
142–
144]. The progression of dental caries was also studied by looking at the microbial composition in white spot lesions, dentin lesions, and deep-dentin lesions [
46]. They saw a continuous shift in the bacterial composition as the caries sites progress through the stages of disease. Together, these works indicate that while the presence of key bacterial species is necessary for caries development, a microbial community is crucial to supporting the persistence of those pathogenic species in the oral cavity. This change appears to be driven by changes in the local environmental conditions, such as a lower pH and changes in the host response. Therefore, monitoring known caries pathogens as well as the rest of the microbial community should provide a global picture of the oral disease state and can act as a tool to predict caries risk and treatment outcomes.
Periodontology
Periodontal diseases range from basic inflammation of the gum tissue to major damage of oral connective tissue and bone. Gingivitis is mild to moderate inflammation of the gum tissue, and diseased tissue can normally be reversed to a healthy state with a proper oral hygiene regimen [
191,
192]. In contrast, periodontitis is diagnosed when inflammation begins to cause permanent damage to the connective tissue surrounding a tooth or the teeth. As the body tries to respond to the presence of bacterial biofilms on the tooth, the host’s immune system actually starts to damage oral tissue. Left untreated, this host-pathogen interaction can lead to loss of gum tissue, loss of attachment of connective tissue to the tooth and bone, and can potentially lead to the loss of teeth [
193]. Extensive research has linked the pathology of gingivitis and periodontitis to certain microbial species such as
Porphyromonas gingivalis,
Treponema denticola,
Tannerella forsythia,
Aggregatibacter actinomycetemcomitans,
Prevotella intermedia/
nigrescens, and
Campylobacter rectus [
146,
147,
194,
195]. These etiological agents are dependent on other members of the oral flora to allow for colonization, and many times there is also an increase in salivary viral load (see later). These bacteria are late colonizers of the biofilm and are not often allowed access to gum tissue when excellent oral hygiene standards are followed.
Reminiscent of dental caries, periodontitis progression is underpinned by a global shift in microbial species [
13,
23,
196]. Extensive work has looked at outlining the composition of the periodontal microbiota in order to help develop more effective treatments and diagnostic testing [
12,
13,
193]. The predicted periodontitis microbiome shift was analyzed using the microarray technology HOMIM to discover the major differences between healthy control patients and those patients that responded well to treatment versus those that suffered from refractory periodontitis [
23]. More species of bacteria in total were detected in diseased patients in comparison to healthy subjects, where 28 % of the 300 species were not detected in healthy individuals. The putative periodontal pathogens were found in much higher numbers than those with healthy periodontium. Those patients with treatable periodontitis were also distinguishable from those with refractory periodontitis based on their unique bacterial profiles. Significantly higher accumulations of common perio-pathogens such as
T. forsythia,
P. gingivalis, and
Prevotella spp. were found within patients with refractory periodontitis compared to treatable but also more “unusual” species that were present, such as
Brevundimonas diminuta,
Mycoplasma salivarium, and
Filifactor alocis [
23]. The microbial profile of the healthy individuals revealed that certain bacterial species were more prevalent in subgingival plaque samples and have been found consistently associated with oral health in other studies, including
Actinomyces spp. [
16,
23],
Capnocytophaga sputigena [
23,
46,
138],
Cardiobacterium hominis [
16,
23],
Haemophilus parainfluenzae [
16,
23],
Rothia dentocariosa/
mucilaginosa [
16,
23],
and Streptococcus sanguinis [
23,
46,
138].
Periodontal pathogens have been found to colonize the tongue and other non-periodontal sites [
38,
197,
198]. The whole saliva provides the immediate source of bacteria for oral biofilm formation and is therefore predicted to have bacterial profiles that are coupled to the biofilm composition.
P. gingivalis,
P. nigrescens T. denticola, and
P. intermedia were found to have a statistical relationship when comparing saliva and periodontal pocket samples [
199]. Additional research has shown both genetic and environmental factors influence bacterial colonization in saliva and the periodontal pocket [
200–
202]. Several groups have assessed the severity of periodontal disease by using levels of both pathogenic bacteria and host-response biomarkers in saliva [
199,
203–
206]. Interestingly, investigations have started to show that the bacterial composition shift paired with host biomarkers is correlated with detecting disease stability or periodontitis disease progression [
27,
204]. Those patients who showed low bacterial and host biomarker levels fared better than those with high bacterial and host biomarker amounts. Kinney et al. [
27] showed that the elevated presence of
Fusobacterium nucleatum,
C. rectus, and
P. intermedia predicted disease progression in 82 % of patients. Low levels of host biomarkers, matrix metalloproteinases 8 and 9 (host-derived enzymes involved with tissue and bone degradation), osteoprotegerin (inhibitor of differentiation and activation of osteoclasts involved in bone resorption), and IL-1 (inflammatory cytokine) predicted periodontal site stability 78 % of the time. These studies present some of the novel work that is starting to link microbiome composition to disease progression and treatment outcomes [
13,
193]. Overall, patients who were successfully treated for periodontitis showed a shift back toward a healthy oral microbiome [
13].
Defining the oral microbiome has allowed researchers to start to understand the normal commensal organisms that are consistent with health. Key work looking at oral-specific disease supported the hypothesis that the oral microbiome usually harbors a core microbiome, and changes in the core composition are directly related to oral disease progression due to dysbiosis. However, it was still unclear if other disease states were linked to the oral microbiome. This shift from homeostasis to dysbiosis reflects a common theme seen across many microbiome profiles that a disease state may not show complete removal and/or enrichment of certain species but instead results in a dramatic shift in the overall proportions of the total microbiome. The microbiome acts like a community that is influenced by the overall presence of key players that may aid in attachment, acquiring nutrients, surviving fluctuations in the environment, etc. Work outlined in the next section will focus on how the oral microbiome has ties to other systemic diseases such as diabetes, autoimmune disorders, hematologic diseases, vitamin deficiencies, and cancer.