Introduction
α-Actinins are myofibril anchor proteins that influence the contractile properties of skeletal muscles. ACTN2 is expressed in slow type I and fast type II fibers, whereas ACTN3 is expressed only in fast fibers. ACTN3 homozygosity for the 577X stop codon (ie, changing 577RR to 577XX, the R577X polymorphism) results in the absence of α-actinin-3 in about 18% of Europeans, diminishes fast contractile ability, enhances endurance performance, and reduces bone mass or bone mineral density. We have examined ACTN3 expression and genetic variation in the masseter muscle of orthognathic surgery patients to determine the genotype associations with malocclusion.
Methods
Clinical information, masseter muscle biopsies, and saliva samples were obtained from 60 subjects. Genotyping for ACTN3 single nucleotide polymorphisms, real-time polymerase chain reaction quantitation of muscle gene message, and muscle morphometric fiber type properties were compared to determine statistical differences between genotype and phenotype.
Results
Muscle mRNA expression level was significantly different for ACTN3 single nucleotide polymorphism genotypes ( P <0.01). The frequency of ACTN3 genotypes was significantly different for the sagittal and vertical classifications of malocclusion, with the clearest association being elevated 577XX genotype in skeletal Class II malocclusion ( P = 0.003). This genotype also resulted in significantly smaller diameters of fast type II fibers in masseter muscles ( P = 0.002).
Conclusion
ACTN3 577XX is overrepresented in subjects with skeletal Class II malocclusion, suggesting a biologic influence during bone growth. ACTN3 577XX is underrepresented in subjects with deepbite malocclusion, suggesting that muscle differences contribute to variations in vertical facial dimensions.
Highlights
- •
The ACTN3 polymorphism R577X associates with skeletal Class II and deepbite malocclusions.
- •
Loss of α-actinin-3 expression changes masseter type II fiber proportions, resulting in deepbite malocclusion.
- •
Two SNPs in ACTN3 at rs1815739 and rs678397 associate Class II malocclusion by altering bone growth.
Malocclusion often develops as a complex trait condition that is influenced by combinations of transcription and growth factors acting on bone, teeth, and skeletal muscles. Complex traits are quantitative or continuous conditions with a broad spectrum of presentations. For humans, variations in height, IQ, blood pressure, and birth weight are complex quantitative traits resulting from the interplay of genetic and environmental influences. One approach for identifying genes that contribute to the development of malocclusion is to consider those already known to influence musculoskeletal growth and function. Malocclusion is a complex musculoskeletal trait because masticatory muscle contributes to variations in the vertical dimension of facial growth. Specifically, vertical facial dimensions are influenced by the size and proportion of muscle fiber types in the masticatory muscles, with the majority of these studies being conducted by direct biopsy or indirect imaging studies of masseter muscles. Genome-wide association analysis of skeletal muscle fiber types is underway and should add important information to the current Human Gene Map for Performance and Health-Related Fitness Phenotypes compendium, which summarizes gene variations that influence muscle size and strength. Overall, these gene association studies demonstrate that fiber type properties are influenced by genetic variations, which most commonly are single nucleotide polymorphisms (SNPs) in gene sequences with functional consequences. Unlike limb muscle, which is highly responsive to training and displays wide phenotypic variability with exercise and other environmental factors, cranial muscles show fewer activity-related changes and are not typically subject to maximum force recruitments.
ACTN3 is a particular gene of interest that influences muscle performance and fiber type proportions. α-Actinins are cytoskeletal proteins that bind actin filaments in a variety of cell types. In skeletal muscle, α-actinin-2 and α-actinin-3 crosslink actin filaments to dense bodies located in the Z disk of the sarcomere, to help order the myofibril array during sarcomere contraction. α-Actinin-2 is found in all skeletal muscle fiber types, whereas α-actinin-3 is restricted to most type II fast-contracting fibers. The genes that encode these 2 closely related isoforms are found on different chromosomes, with ACTN2 located on the long arm of chromosome 1 and ACTN3 on chromosome 11. A common nonsense mutation R577X identified in the ACTN3 gene results in a lack of protein expression because of the production of a stop codon at residue 577. About 18% of the European population is homozygous for the R577X change. Absence of α-actinin-3 is not associated with any obvious pathology, and since α-actinin-2 is still expressed in the fast fiber types, the functional role for the α-actinin-3 was first thought to be redundant. Shortly thereafter, it became apparent that ACTN3 genotype variations are important in human elite athletic performances. In a study comparing Australian Olympic athletes with controls, both male and female elite sprint athletes had higher frequencies of the 577R allele. Among females, elite sprint athletes also had a higher 577RX heterozygote frequency, and elite endurance athletes had a lower 577RX frequency. There was no comparable heterozygote genotype effect in male athletes. Subsequently, ACTN3 allele and genotype frequencies have been investigated in at least 10 other athletic and control populations. These investigations support the conclusion that the 577RR genotype is more common in sprint and power athletes, but not that the X allele enhances endurance capability. Overall, the literature indicates that α-actinin-3 enhances the production of forceful, fast contraction in type II muscle fibers and that these genotypic effects may be influenced by sex.
α-Actinin-3 may also contribute to variations in muscle function by interaction with the signaling protein calcineurin to influence fiber type proportions during growth. α-Actinin-3 binds to the calsarcin family signaling proteins located at the Z disc ; that in turn binds to calcineurin to activate fiber type specific gene expression pathways that determine fiber types and size. In a study of vastus lateralis muscles in young men, the RR genotype resulted in type IIX, fast-contracting fatigable fibers, of larger size and greater number compared with the XX genotype. Consequently, men with the RR genotype had significantly elevated leg muscle power. Type I, slow-contracting fibers also show variations with ACTN3 genotype in vastus lateralis, with the percentage of type I fibers increasing in 577XX compared with the RR genotype. Therefore, α-actinin-3 may act directly through structural functioning or cell-signaling pathways to alter the composition and function of skeletal muscles.
Previous masseter muscle biopsy studies have demonstrated that increased sizes or proportions of type II fibers are associated with skeletal deepbite malocclusion, and decreased type II fibers are associated with skeletal open bite. To further explore how genetic variations might influence masticatory muscle function and skeletal shape, in this study, we sought to associate ACTN3 genotypes with malocclusion classification and masseter muscle fiber type properties. To do so, we looked at 2 SNPs located at rs1815739 and rs678397. The first SNP, rs1815739 (R577X), is a cytosine to thymine transition at nucleotide 1,586 in exon 16 that converts an arginine to a stop codon at residue 577 and produces 3 genotypes: CC (normal), TC (heterozygote), and TT (no α-actinin-3). The second SNP, rs678397, is a cytosine to thymine transition at nucleotide 15,193 in an ACTN3 gene intron, which has no reported functional changes, and produces 3 genotypes—CC (normal), TC, and TT.
Material and methods
Sixty subjects undergoing orthodontic and maxillofacial surgery treatment for correction of malocclusion were recruited from the Department of Oral and Maxillofacial Surgery at the University of Lille in France. They were recruited after they had signed an informed consent, and the research protocol was validated by the French independent ethical committee and the Temple University and the University of Pittsburgh institutional review boards. Malocclusion classification was based on the sagittal and vertical jaw repositioning required to execute the surgical treatment plan. The subjects were classified into 1 of 6 craniofacial morphologic groups that included 1 variation of sagittal skeletal jaw malocclusion, either Class II or Class III, and 1 variation of vertical skeletal jaw malocclusion—open bite, deepbite, or normal bite—relationship. Thirty-one untreated Class I control subjects without malocclusion came from the Dental Registry and DNA Repository at the University of Pittsburgh; they included only subjects of European descent, to be comparable with the French subjects with malocclusion. Saliva samples from all subjects were stored in Oragene kits (DNA Genotek, Ottawa, Ontario, Canada) and used for DNA extraction and posterior genotyping. For the malocclusion subjects, muscle samples were also collected from the deep anterior area of the masseter muscles as the bilateral sagittal split osteotomy of the mandible was performed. After snap freezing, muscle samples were stored at –80°C before histologic and gene expression analysis.
Two SNPs in ACTN3 were selected for genotyping on all subjects—rs1815739 (the R577X SNP) and rs678397 (a SNP not known to have functional consequences)—and tested to determine whether specific allelic variants are overrepresented in subjects with malocclusion subclassifications using TaqMan chemistry (Life Technologies, Carlsbad, Calif) and end-point analysis in an automatic sequence-detection instrument (ABI Prism 7900HT; Applied Biosystems, Foster City, Calif), as described previously. Thirty-three additional anonymous SNPs were genotyped to assess the presence of population substructure among the controls.
We performed fiber type histomorphometric analysis of the masseter muscles. Frozen muscles were cryosectioned at 10-μm thickness to obtain serial cross-sectional slices, and sections were mounted on glass microscope slides for immunostaining with 5 antibodies specific for myosin heavy chain (MyHC) isoforms—antitype I, antitype II, antitype IIA, antitype neonatal, and anti-α-cardiac (atrial)—as described previously. We classified masseter fibers into 4 fiber type groups as type I, type hybrid (containing both types I and II MyHCs), type II containing only type IIA and/or IIX MyHCs, and type neonatal-atrial that contained the neonatal and/or α–cardiac MyHCs in combination with other type I and II isoforms. Type I fibers are slow contracting and fatigue resistant, used most commonly to maintain postural freeway space. Type II fibers are fast contracting and are either fatigue resistant (type IIA) or fatigable (type IIX). The hybrid fibers are an unusual and distinctive fiber type found in masseter muscles; they combine slow and fast contractile properties. Hybrid fibers are more commonly found in certain states of skeletal muscle pathology, although they are sometimes present in normal healthy limb muscles. In human masticatory muscles, however, hybrid fibers are always present as a common fiber type. In addition, α-actinin-3 was identified across fiber types by staining with a rabbit IgG monoclonal antibody (EP2531Y), commercially available from OriGene Technologies (Rockville, Md). For fiber type classification, only tissue section series with consistent antibody reactions for all stains and acceptable morphology of muscle fibers, which were clearly in transverse section, were used. All fibers in the selected areas were classified by type, and their cross-sectional areas were measured with ImageJ image analysis software (National Institutes of Health, Bethesda, Md). Tests for measurement error included intrarater reliability in determination of fiber area (by repeating morphometric tracing of all fiber areas in 1 biopsy by 1 examiner [J.J.S.]); this resulted in an R 2 value of 0.94. We compared differences in fiber type properties between all malocclusion subjects who were genotyped as either 577XX or RR.
RNA was isolated from the remaining muscle after cryosectioning with TRIzol reagent (Life Technologies) and quantified by absorbance at A 260 as described previously. ACTN2 and ACTN3 were quantified by TaqMan quantitative real-time polymerase chain reaction. Reactions, in triplicate, contained RNA from masseter muscle or an adult skeletal muscle reference standard, TaqMan RNA-to-CT 1-step reagent, and an Applied Biosystems expression assay set for ACTN2 (#Hs01100111_g1), ACTN3 (#Hs00153809_m1), or internal control HPRT1 (hypoxanthine phosphoribosyltransferase 1) (#Hs02800695_m1) for normalization. RNA was expressed as relative quantities determined by the comparative cycle threshold method that measures fold differences between normalized quantities of the target in the sample and the reference standard. Thirteen subjects were included in the mRNA quantification to determine whether there were differences in message expression between ACTN3 genotypes rs1815739 and rs678397.
Statistical analysis
Chi-square or Fisher exact tests were used to assess Hardy-Weinberg equilibrium and compare allele and genotype distributions between the subjects with malocclusion and the untreated Class I control subjects. Student t tests were used to compare differences for mean fiber type areas, fiber numbers, and percent occupancy measurements between the ACTN3 genotypes. Analysis of variance (ANOVA) was used to compare differences in malocclusion classifications for the ACTN3 genotypes between the malocclusion and control subjects. A separate ANOVA was conducted to determine whether ACTN2 and 3 mRNA expression levels were different by R577X genotype.
Statistical analysis
Chi-square or Fisher exact tests were used to assess Hardy-Weinberg equilibrium and compare allele and genotype distributions between the subjects with malocclusion and the untreated Class I control subjects. Student t tests were used to compare differences for mean fiber type areas, fiber numbers, and percent occupancy measurements between the ACTN3 genotypes. Analysis of variance (ANOVA) was used to compare differences in malocclusion classifications for the ACTN3 genotypes between the malocclusion and control subjects. A separate ANOVA was conducted to determine whether ACTN2 and 3 mRNA expression levels were different by R577X genotype.
Results
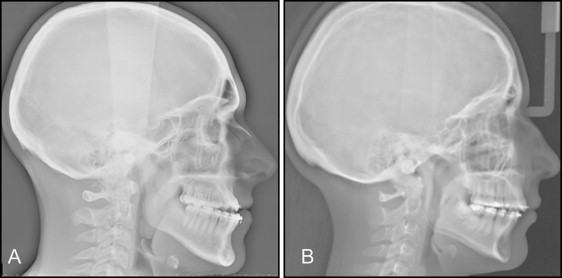
For the ACTN3 genotype, we recruited 60 orthognathic surgery patients who were systemically healthy and without genetic craniofacial syndromes, other growth disturbances, or reported trauma. They were 42 women and 18 men, with the average age of 23.5 + 9.9 years, including 18 with Class II open bite, 7 with Class III open bite, 13 with Class II deepbite, 6 with Class III deepbite, 12 with Class II normal bite, and 4 with Class III normal bite malocclusions. R577X genotype frequencies for the entire malocclusion group were 14% CC, 63% TC, and 23% TT, with the reported European population frequency of 18% for TT. Lateral cephalograms of 1 Class III open-bite subject with R577X CC genotype and 1 Class II open-bite subject with TT genotype are shown in Figure 1 . Because we hypothesized that the ACTN3 SNPs would differ between the sagittal and vertical malocclusion classifications, we compared SNP frequency for Class II vs Class III with the Class I controls ( Table I ), and between those with open bite, normal bite, and deepbite ( Table II ). Overall, only a small number of genotypes were undetermined, ranging from 0 to 3 for each comparison.
| SNP marker | Phenotype | Genotypes | Total (n) | P value | |||||
|---|---|---|---|---|---|---|---|---|---|
| CC | TC | TT | Undetermined | n | Genotypes | Alleles | |||
| ACTN3 rs1815739 | Class III | 4 (25%) | 9 (56%) | 3 (19%) | 1 | 17 | 0.39 | 0.28 | |
| Class II | 4 (10%) | 27 (66%) | 10 (24%) | 2 | 43 | 60 | 0.003 ∗ | 0.009 ∗ | |
| Control | Class I | 14 (45%) | 12 (39%) | 5 (16%) | 31 | ||||
| ACTN3 rs678397 | Class III | 4 | 9 | 3 | 1 | 17 | 0.11 | 0.04 | |
| Class II | 4 (10%) | 24 (60%) | 12 (30%) | 3 | 43 | 60 | 0.003 ∗ | 0.000005 ∗ | |
| Control | Class I | 17 (55%) | 12 (39%) | 2 (6%) | 31 | ||||
Stay updated, free dental videos. Join our Telegram channel

VIDEdental - Online dental courses


