Fig. 4.1
Long-term implant survival decreased in patients with a history of severe periodontitis (From Levin et al. 2011)
Reviews of these clinical studies highlight that patients with no history of periodontitis demonstrated a better outcome of long-term stability in implant treatment and that the failure rate in patients with the history of periodontitis tended to be more prominent at the prosthetic phase (Klokkevold and Han 2007; Ong et al. 2008). These studies did not provide the possible cause of implant failure and the qualitative nature of peri-implant tissue evaluations were often less informative. However, it may be postulated that a combination of genetic and environmental factors that had initially caused severe periodontitis could later contribute to the implant failure.
4.3.2 Quantity and Quality of Alveolar Bone
The extensive review of published data by Esposito et al. (1998) suggested a distinct trend of higher failure prevalence in implants placed in the maxillary molar region (Esposito et al. 1998). This may not be simply due to the factors associated with a maxillary location (Eckert and Wollan 1998) but rather due to the quantity and/or quality of bone tissue at the maxillary molar site of implant placement, which may be attributed to one of the etiologic factors for dental implant failure (Jaffin and Berman 1991). Leckholm and Zarb (1985) proposed an index describing the operators’ subjective assessment of alveolar bone quantity and quality (Lekholm and Zarb 1985) (Fig. 4.2).
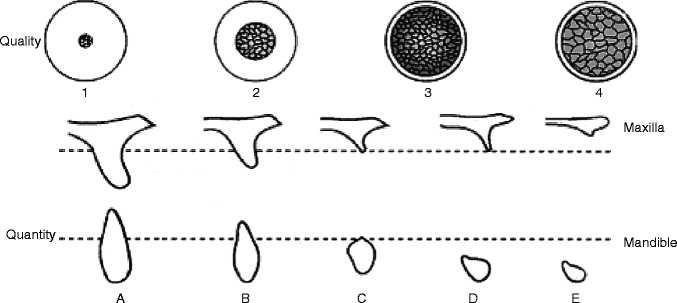

Fig. 4.2
Bone quality classification (Leckholm and Zarb) and bone quantity classification (Atwood)
Clinically, the maxillary posterior residual ridge was often classified as type 4 bone, depicting the thin cortical bone thickness with a less developed trabecular pattern. Therefore, it has been postulated that poor quality of alveolar residual ridge is a risk factor. In a retrospective cohort study of 4,680 implants over a 21-year period, Moy et al. (2005) reported the poor bone quality to be one of the associated factors for the increased implant failure (Moy et al. 2005).
In a retrospective study, Hardt et al. (2002) reported that patients with a history of periodontitis appeared to develop poorer bone quantity (index D) but not bone quality (Hardt et al. 2002). The biological parameters contributing to the poor bone quantity and quality of alveolar residual ridge are still debated (Jahangiri et al. 1998; Kingsmill 1999). Nonetheless, it is highly conceivable that the genetic and environmental factors that contributed to the loss of quantity and quality of alveolar bone may also affect the long-term stability of implant osseointegration.
4.4 Implant Failure and Periodontitis-Associated Genes
With the advent of the human genome project and the successful completion of the entire genetic sequence of individuals with different ethnic backgrounds, the sequence mismatch between those individuals has generated a new concept of genetic contributions to common disease. Genetic diseases caused by genetic mutations such as craniosynostosis syndromes occur in less than 1 % of a given population by definition. On the contrary, genetic variations resulting from the gene sequence mismatch between individuals can occur more frequently, and thus, single gene sequence mismatch should not cause serious problems in an ordinary environment. However, when seemingly unrelated genetic variations are combined, the individual may develop a so-called common disease. It is also postulated that the combination of genetic variations and environmental conditions may collectively manifest the increased susceptibility.
Variations in the DNA sequences are small in humans; however, they have been shown to cause diseases; to respond differently to pathogens, drugs or vaccines; and to determine the success and failure of certain treatments. A single-nucleotide polymorphism (SNP) presents the most frequent sequence variation (Fig. 4.3). SNP occurs approximately 1 in every 300 base pairs (bp) of human genome, which contains three billion bp. Along with the completion of more genetic sequences, increasing numbers of SNPs are updated in the publically available databases.


Fig. 4.3
Single-nucleotide polymorphism (SNP) presents the most frequent genomic variations. Some SNPs were found to modify the encoding amino acids. For example, TAC encodes for tyrosine. When T is substituted by C, CAC now encodes histidine. This type of SNP is called a non-synonymous SNP. Other SNPs such as GGT vs. GGC do not change the encoding amino acid (glycine), which is called a synonymous SNP
Many SNPs are found in protein non-coding domains or between active genes. Therefore, it is highly conceivable that these SNPs do not cause any serious problems. However, SNPs that are found in the protein-coding region may present profound manifestations. An amino acid unit of a protein is encoded by three base pairs (bp) of DNA, and an SNP within this region may result in an amino acid substitution (non-synonymous SNP). As a result, the variant of protein sequence may lead to an altered structural and functional property potentially causing a significant effect. Among all SNPs, however, non-synonymous SNPs form a small fraction.
Synonymous SNPs are also found to occur within the protein-coding domain called the exon in chromosomal DNA. This type of SNP does not contribute to any modification in the protein sequence and thus is considered a “silent” SNP. The majority of SNPs are found in the non-coding domains such as introns, promoter regions and 3’ regions. The putative effect of these SNPs has not been fully elucidated. It is possible that the function of chromosomal DNA, such as the rate of gene transcription, may be affected by SNPs located in the promoter region.
When the genetic material is inherited by offspring, parental DNAs are not admixed randomly but rather transferred as large sections. There is a strong tendency that even after many generations, these large sections are maintained in the chromosomal DNA of the offspring. The surviving large sections, called the “haplotype,” may contain the protein-coding sequences and their adjacent DNA sequences. Therefore, SNPs within the haplotype should show significant homology between the family members and to a large extent within the same ethnic groups. These SNP haplotypes provide useful information for elucidating the inheritance of a certain trait.
A simple oral rinse gives rise to an ample amount of chromosomal DNA from the patient, which may be applied for polymerase chain reaction (PCR) analysis of target SNPs. Statistical association between SNPs in the target genes and patients’ phenotype (implant failure) has established a new platform of investigation in dentistry (Nishimura and Garrett 2004).
4.4.1 Interleukin-1 (IL-1)-Related Genes
Overzealous immunological reactions are believed to cause abnormal tissue destruction leading to periodontitis. Inflammatory cytokines are involved in the initiation and continuation of such immune reactions. Interleukin-1 (IL-1) has been long recognized in periodontal tissues and its relevance to the development of periodontitis has been established (Page 1991). IL-1 receptor recognizes both IL-1 α and IL-1β, which are encoded by distinct genes but share approximately 50 % sequence homology. IL-1 receptor also binds to IL-1 receptor antagonists (IL-1RA, encoded by IL-1RN gene). Whereas IL-1α or IL-2β exerts downstream reactions after binding to IL-1 receptor, IL-1RA simply occupies the IL-1 receptor without any intracellular effects and thus is considered an inhibitor of IL-1 functions (Arend and Guthridge 2000). Among these IL-1-related molecules, it has been reported there is an increased level of IL-1β in gingival crevicular fluid associated with severe periodontitis (Chaudhari et al. 2011; Honig et al. 1989; Liu et al. 1996) and peri-implantitis (Curtis et al. 1997; Kao et al. 1995; Panagakos et al. 1996; Tsai et al. 1995).
Kornman et al. (1997) reported that the genotype exhibiting concurrent minor allele SNPs in IL-1α (−889) and IL-1β (3953) were found to be more prevalent in non-smoker patients with severe chronic periodontitis (Kornman et al. 1997). Initially, the assay for IL-1α (−889) SNP presented some technical difficulties. However, IL-1α (−889) SNP was later found within the same “haplotype” of IL-1α (4845) SNP. When we inherit the genetic information from our ancestors, a block of chromosome often containing multiple genes (called alleles) is transmitted as one piece. This piece of chromosomal fragment is called haplotype. In other words, the IL-1α (−889) SNP located before the beginning of the IL-1α coding sequence and IL-1α (4845) SNP located the end of IL-1α coding sequence should be transmitted together over many generations. Therefore, the presence of IL-1α (−889) SNP should be predicted by the test for IL-1α (4845) SNP, which provided a technically feasible assay. Currently, SNPs of IL-1α (−889) and IL-1α (4845) are interchangeably used. It was also found that due to early sequence irregularities, IL-1β (3953) SNP may be interchangeably described as IL-1β (3954).
A commercially available clinical test of IL-1 SNPs was developed to determine the SNP minor alleles of IL-1α (−889/4845) and IL-1β (3953/3954). PTS® genetic susceptibility test is the only commercially available SNP test in dentistry and currently distributed by Interleukin Genetics, Inc. and by OralDNA Labs (limited to the US market). In this test, patients who carry the minor allele of both IL-1 SNP sites would be informed of a potentially increased susceptibility to severe chronic periodontitis. The availability of the PTS® test may have contributed to the robust increase in genetic association studies for both periodontitis and implant failure (Table 4.1).
Table 4.1
SNP genotypes of inflammation-related genes and implant failure
|
Gene
|
SNP
|
Association
|
Implant failure phenotypes
|
Smoking variant
|
Implant system
|
Reference
|
|---|---|---|---|---|---|---|
|
IL1A
|
−889
|
None
|
Mobility, pain in pre-prosthetic stages
|
Non-smokers
|
3I
|
Campos et al. (2005a)
|
|
−889
|
None
|
Biological complications
|
Mixed
|
Branemark System
|
Jansson et al. (2005)
|
|
|
−889
|
None
|
Implant failure
|
Rogers et al. (2002)
|
|||
|
−889
|
None
|
Implant loss/biological complications
|
Mixed
|
Vaz et al. (2011)
|
||
|
−889
|
None
|
Lack of osseointegration
|
Mixed
|
ITI
|
De Boever and De Boever (2006)
|
|
|
−889
|
None
|
Marginal bone loss at stage II surgery
|
Astra Tech
|
Shimpuku et al. (2003)
|
||
|
−889
|
None
|
Marginal bone loss >3 threads
|
Mixed
|
Branemark System
|
Laine et al. (2006)
|
|
|
−889
|
None
|
Marginal bone loss at stage II surgery
|
Mixed
|
Lin et al. (2007)
|
||
|
4845
|
p < 0.04
|
Marginal bone loss/year (protective?)
|
Heavy smokers
|
ITI
|
Feloutzis et al. (2003)
|
|
|
4845
|
p = 0.0022
|
Biological complications
|
Smokers
|
ITI
|
Gruica et al. (2004)
|
|
|
IL1B
|
−511
|
p = 0.083
|
Loss of multiple implants
|
Mixed
|
NEODENT
|
Dirschnabel et al. (2011)
|
|
−511
|
p = 0.033
|
Marginal bone loss at stage II surgery
|
Astra Tech
|
Shimpuku et al. (2003)
|
||
|
−551
|
p = 0.021
|
Marginal bone loss at stage II surgery
|
Mixed
|
Lin et al. (2007)
|
||
|
−511
|
None
|
Mobility, pain in pre-prosthetic stages
|
Non-smokers
|
3I
|
Campos et al. (2005b)
|
|
|
−511
|
None
|
Peri-implantitis
|
Melo et al. (2011)
|
|||
|
−511
|
None
|
Marginal bone loss >3 threads
|
Mixed
|
Branemark System
|
Laine et al., (2006)
|
|
|
3953
|
None
|
Mobility, pain in pre-prosthetic stages
|
Non-smokers
|
3I
|
Campos et al. (2005a)
|
|
|
3953
|
None
|
Biological complications
|
Mixed
|
Branemark System
|
Jansson et al. (2005)
|
|
|
3953
|
None
|
Implant failure
|
Rogers et al. (2002)
|
|||
|
3953
|
None
|
Implant loss/biological complications
|
Mixed
|
Vaz et al. (2011)
|
||
|
3953
|
None
|
Lack of osseointegration
|
Mixed
|
ITI
|
De Boever and De Boever (2006)
|
|
|
3954
|
p < 0.04
|
Marginal bone loss/year (protective?)
|
Heavy smokers
|
ITI
|
Feloutzis et al. (2003)
|
|
|
3954
|
p = 0.0022
|
Biological complications
|
Smokers
|
ITI
|
Gruica et al. (2004)
|
|
|
3954
|
None
|
Biological complication
|
Mixed
|
NEODENT
|
Montes et al. (2009)
|
|
|
3954
|
None
|
Peri-implantitis
|
Melo et al. (2011)
|
|||
|
3954
|
None
|
Marginal bone loss at stage II surgery
|
Astra Tech
|
Shimpuku et al. (2003)
|
||
|
3954
|
None
|
Marginal bone loss >3 threads
|
Mixed
|
Branemark System
|
Laine et al. (2006)
|
|
|
3954
|
None
|
Marginal bone loss at stage II surgery
|
Mixed
|
Lin et al. (2007)
|
||
|
IL-RN
|
Intron 2
|
p = 0.029
|
Loss of multiple implants
|
Mixed
|
NEODENT
|
Montes et al. (2009)
|
|
Intron 2
|
p = 0.02
|
Marginal bone loss >3 threads
|
Branemark System
|
Laine et al. (2006)
|
||
|
Intron 2
|
None
|
Mobility, pain in pre-prosthetic stages
|
Non-smokers
|
3I
|
Campos et al. (2005b)
|
|
|
IL2
|
−330
|
None
|
Mobility, pain in pre-prosthetic stages
|
Branemark System
|
Campos et al. (2005a)
|
|
|
IL6
|
−174
|
None
|
Peri-implantitis
|
Melo et al. (2011)
|
||
|
−174
|
None
|
Mobility, pain in pre-prosthetic stages
|
Branemark System
|
Campos et al. (2005a)
|
||
|
IL10
|
Promoter
|
None
|
Implant loss
|
NEODENT
|
Pigossi et al. (2012)
|
|
|
TNFa
|
−308
|
None
|
Mobility, pain in pre-prosthetic stages
|
3I
|
Campos et al. (2004)
|
|
|
−308
|
None
|
Peri-implantitis
|
Non-smokers
|
Cury et al. (2009)
|
||
|
TGFb
|
−509
|
None
|
Mobility, pain in pre-prosthetic stages
|
3I
|
Dos Santos et al. (2004)
|
|
|
−800
|
None
|
Mobility, pain in pre-prosthetic stages
|
3I
|
Dos Santos et al. (2004)
|
||
|
MMP1
|
−1607
|
p < 0.001
|
Mobility, pain in pre-prosthetic stages
|
3I
|
Santos et al. (2004)
|
|
|
−1607
|
p = 0.046
|
Loss of implant during healing stage
|
Non-smokers
|
Arisan et al. (2005)
|
||
|
−1607
|
p = 0.011
|
Mobility, pain in pre-prosthetic stages
|
Leite et al. (2008)
|
|||
|
−516
|
None
|
Mobility, pain in pre-prosthetic stages
|
Leite et al. (2008)
|
|||
|
MMP9
|
−1562
|
None
|
Mobility, pain in pre-prosthetic stages
|
3I
|
Santos et al. (2004)
|
|
Feloutzis et al. (2003) investigated 180 patients, who received at least one ITI implant at the University of Berne (Feloutzis et al. 2003). In this study, 7 out of a total of 182 implants were lost during the loading period, all of which were placed in the maxilla of heavy smokers defined as 20 cigarettes per day for at least 20 years. For the remaining implants, the periapical radiographs taken during the multiple recall visits were used to measure the average loss of bone heights mesial and distal to the implant surface (the absolute bone loss, ABL). ABL as well as the bone loss per year was found to be significantly greater in the heavy smoker group; however, these peri-implant bone loss values were not associated with an IL-1 genotype based on the PTS® test (Feloutzis et al. 2003). Contrary to the hypothesis, there was a general but insignificant trend that IL-1 genotype positive patients showed smaller bone loss values as well as less bleeding-on-probing sites. When the smoking habits and IL-1 genotypes were combined, non-smokers or former smokers carrying the positive IL-1 genotypes indicated significantly less peri-implant bone loss (ABL and the bone loss per year). The smokers with the positive and negative IL-1 genotypes showed similar bone loss values. Therefore, if patients do not smoke, the data from this study suggest that the positive IL-1 genotype by PTS® test may indicate protection from the peri-implant bone loss.
Gruica et al. (2004) reported a follow-up study using the increased number of patients in the same institution (Gruica et al. 2004). This study included 180 patients and peri-implant biological complications were found associated with the longer number of years smoked (p = 0.0101) and the greater number of implants placed (p = 0.0075). On the contrary, IL-1 minor allele genotypes examined by PTS® test failed to show any correlation with peri-implant biological complications. However, detailed examinations revealed that 12 patients in this study were current heavy smokers carrying the IL-1 minor allele genotypes and 6 patients in this group exhibited biological complications. Periapical radiographs taken at recall visits of 1 and 8 years and radiographic peri-implant bone loss values were determined during this period of 7 years. The statistical evaluation using a generalized estimating equation suggested a correlation between IL-1 minor allele genotypes and peri-implant bone loss.
The IL-1 genotypes have been repeatedly examined in patients who have experienced implant failure or peri-implantitis. However, the results from all authors failed to establish the statistically significant correlation between IL-1α (−889/4845) and IL-1β (3953/3954) dominant minor SNP alleles, with various biological complications associated with dental implants (Table 4.1) (Campos et al. 2005b; Jansson et al. 2005; Laine et al. 2006; Lin et al. 2007; Montes et al. 2009; Rogers et al. 2002; Shimpuku et al. 2003; Wilson and Nunn 1999). As such, the IL-1α (−889/4845) and IL-1β (3953/3954) SNP genotype alone may not explain the cause of implants failure (Bormann et al. 2010).
Several investigations examined different SNP genotypes associated with IL-1 genes such as IL-1β (−511) or IL-1RN (intron 2) and found suggestive or strong correlations with the loss of multiple implants or marginal bone loss (Dirschnabel et al. 2011; Laine et al. 2006; Lin et al. 2007; Montes et al. 2009; Shimpuku et al. 2003).
In a recent study, while statistical significance was not achieved, there was an increased trend of the prevalence of these SNP minor allele genotypes in patients experiencing peri-implantitis among the other evaluated implant-related biological complications (Vaz et al. 2011). Rogers et al. (2002) reported no significant correlation between the IL-1α (−889/4845) and IL-1β (3953/3954) SNP genotype and implant failure; however, severe adult chronic periodontitis was significantly associated with IL-1β (3953/3954) SNP, but not IL-1α (−889/4845) (Rogers et al. 2002).
The IL-1-related genes are located in close proximity within the approximately 300 Kb region chromosome 2q14 (Fig. 4.4a). The number of publications on the positive association of IL-1 genes with implant failure appeared to increase in the area between IL-1β (−511) and IL-RN (intron 2) (Fig. 4.4b). Incidentally, an analysis of this IL-1 cluster region comparing 40 generalized aggressive periodontitis patients and 96 periodontally healthy subjects similarly reported the elevated association between IL-1β (−511) and IL-RN (intron 2) (Fig. 4.4c) (Scapoli et al. 2005).
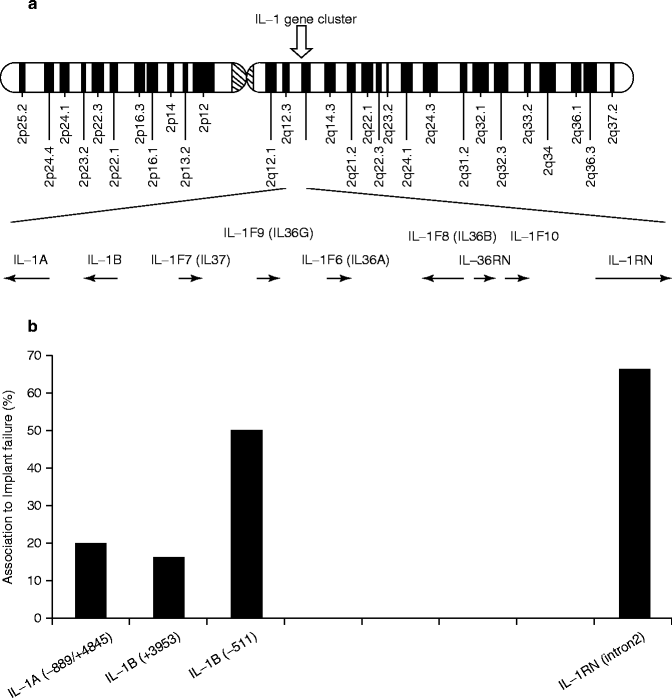
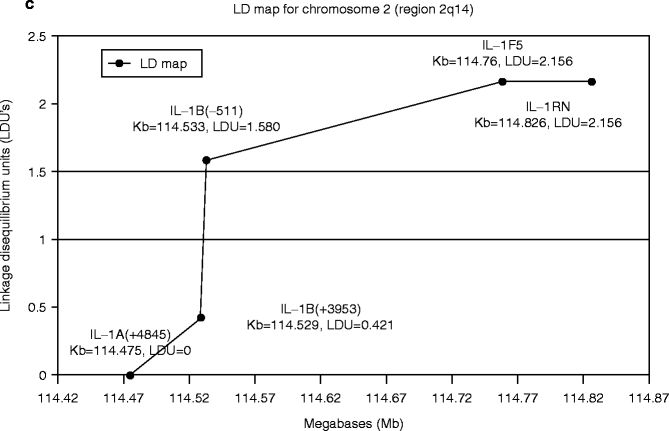


Fig. 4.4
(a) Diagram of human chromosome 2. The IL-1-related genes are located in close proximity within the approximately 300 Kb region chromosome 2q14. (b). The number of published reports on the positive association between implant failure and IL-1 cluster gene SNPs. (c) Linkage disequilibrium map of IL-1 cluster region for aggressive periodontitis (From Scapoli et al. 2005)
Melo et al. (2011) did not find any differences in the genotypic variations in IL-1β (3953/3954), IL-1β (−511) and IL-6 (−174) SNPs in peri-implantitis patients, and the same patient groups did not show the increase or decrease in the concentrations of IL-1b and IL-6 in the crevicular fluid sample obtained from healthy osseointegrated implants, peri-implantitis sites as well as healthy teeth (Melo et al. 2011). However, this study did not evaluate IL-1RN and IL-36RN.
Stay updated, free dental videos. Join our Telegram channel

VIDEdental - Online dental courses


