Chapter 5 Dental plaque
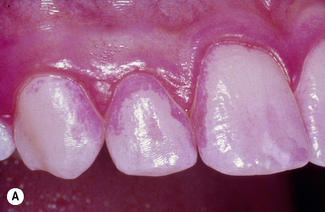
Dental plaque is a general term for the complex microbial community that develops on the tooth surface, embedded in a matrix of polymers of bacterial and salivary origin. Plaque that becomes calcified is referred to as calculus or tartar. The presence of plaque in the mouth can readily be demonstrated by rinsing with a disclosing solution such as erythrosin (Fig. 5.1). The majority of plaque is found associated with the protected and stagnant regions of the tooth surface such as fissures, approximal regions between teeth, and the gingival crevice (see Figs 2.2 and 5.1). Plaque is found naturally on the tooth surface, and forms part of the host defences by excluding exogenous (and often pathogenic) species (colonization resistance) (Table 4.7). On occasions, however, plaque can accumulate beyond levels compatible with oral health, and this can lead to shifts in the composition of the microflora and predispose sites to disease (Ch. 6). Dental plaque is an example of a biofilm and a microbial community, and the significance of this will be explained in the following sections.
Microbial biofilms
Numerous studies of a range of distinct ecosystems have shown that the vast majority of microorganisms exist in nature associated with a surface. The term ‘biofilm’ is used to describe communities of microorganisms attached to a surface; such microbes are usually spatially organized into a three-dimensional structure and are enclosed in a matrix of extracellular material (sometimes termed a ‘glycocalyx’) derived both from the cells themselves and from their environment. If (a) biofilm microbes were simply planktonic (liquid-phase) cells that had adhered to a surface, and (b) the properties of microbial communities were merely the sum of those of the constitutive populations, then scientific interest in such issues would be limited. However, research over recent years has revealed that cells growing as biofilms have unique properties, some of which are of clinical significance (Table 5.1), for example, biofilms can be up to 1000 times more tolerant of antimicrobial agents than the same cells growing in liquid culture, while communities of interacting species can be more pathogenic than pure cultures of the constituent microorganisms.
Table 5.1 General properties of biofilms and microbial communities.
| General property | Dental plaque example |
|---|---|
| Open architecture | presence of channels and voids |
| Protection from host defences, desiccation, etc | production of extracellular polymers to form a functional matrix; physical protection from phagocytosis |
| Enhanced tolerance to antimicrobials* | reduced sensitivity to chlorhexidine and antibiotics; gene transfer |
| Neutralization of inhibitors | β-lactamase production by neighbouring cells to protect sensitive organisms |
| Novel gene expression* | synthesis of novel proteins on attachment; up-regulation of gtfBC in mature biofilms |
| Coordinated gene responses | production of cell-cell signalling molecules (e.g. CSP, AI-2) |
| Spatial & environmental heterogeneity | pH & O2 gradients; coadhesion |
| Broader habitat range | obligate anaerobes in an overtly aerobic environment |
| More efficient metabolism | complete catabolism of complex host macromolecules (e.g. mucins) by microbial consortia |
| Enhanced virulence | pathogenic synergism in abscesses and periodontal diseases |
* one consequence of altered gene expression can also be an increased tolerance of antimicrobial agents.
Originally, biofilms were considered to be dense, compressed accumulations of cells, and this compacted structure was believed to be responsible for many of the novel properties of biofilms. Recent advances in the study of biofilms have come from the application of novel techniques that enable biofilms to be studied in situ without any processing of samples that could distort their structure. A prime example has been the use of confocal laser scanning microscopy (CLSM) to study biofilm architecture without chemical fixation or embedding techniques. Optical thin sections can be generated throughout the depth of the biofilm, and these can be combined using imaging software to generate three-dimensional images. In addition, the location of specific organisms can be visualized with immunological or oligonucleotide probes, such as by fluorescent in situ hybridization, FISH, (Fig. 3.3), while other molecular probes can indicate the vitality and metabolic activity of cells. CLSM can also be used in combination with ‘reporter genes’ to identify genes that are expressed only when cells form a biofilm. This technology involves the insertion of a marker into the bacterial chromosome downstream of a promoter so that a recognisable ‘signal’ (e.g. fluorescence) is produced when the gene is activated.
The application of these modern approaches to the general field of microbial ecology has shown that biofilms that develop in low-nutrient environments, especially those from aquatic habitats, have a more open structure than had been predicted previously from earlier studies employing electron microscopy. Channels have been observed in biofilms of environmental bacteria, enabling potentially growth-limiting factors such as nutrients and oxygen to penetrate more extensively than previously thought. In addition, the use of micro-electrodes and chemical probes have shown that considerable gradients in key factors (pH, redox potential, etc.) can occur over relatively short distances (a few microns, i.e. a few cell diameters) within biofilms. This produces spatial and temporal heterogeneity within the biofilm enabling fastidious bacteria to survive in apparently hostile or incompatible environments (Table 5.1).
Biofilm bacteria are phenotypically distinct from planktonic cells, and one particularly important aspect of this is an increased tolerance of biofilm cells to antimicrobial agents. An extreme example was the finding that P. aeruginosa growing on urinary catheter material was between 500–1000 times more tolerant of the antibiotic, tobramycin, than the same cells in liquid (planktonic) culture. Conventionally, the sensitivity of bacteria to antimicrobial agents is determined on cells grown in liquid culture by the measurement of the minimum inhibitory concentration (MIC) or minimum bacteriocidal concentration (MBC) against relevant antimicrobial agents. Given the decreased sensitivity of an organism on a surface to antimicrobial agents, it has been argued that it would be more appropriate to determine the ‘biofilm inhibitory concentration (BIC)’ and ‘biofilm killing concentration (BKC)’ or ‘biofilm eradicating concentration (BEC)’. As yet, these proposals have not been widely accepted, and there are no generally agreed methods by which these concentrations could be determined.
Biofilms in the mouth
Dental plaque is one of the best studied biofilms, and displays all of the characteristic features of a typical biofilm (Table 5.1). Compared to other habitats, dental plaque is relatively accessible for sampling, and can even be grown on relevant removable surfaces for subsequent investigation and experimentation in the laboratory (in situ models; Ch. 4). A large proportion of the key microorganisms can be identified, while the phenotype of such microbes (adhesins, metabolic potential, cell–cell interactions) is well-characterized. Extensive biofilms also develop on dentures, and the tongue. On other mucosal surfaces, bacteria attach to epithelial cells and may develop a surface-associated phenotype, but extensive 3-dimensional biofilms do not usually develop. Another dentally-relevant biofilm develops on the tubing used in dental unit water supply systems (DUWS); this will be discussed in Chapter 12 (see Fig. 12.6), but the principles governing the formation and properties of these biofilms will be similar to those described below for dental plaque.
Mechanisms of dental plaque formation
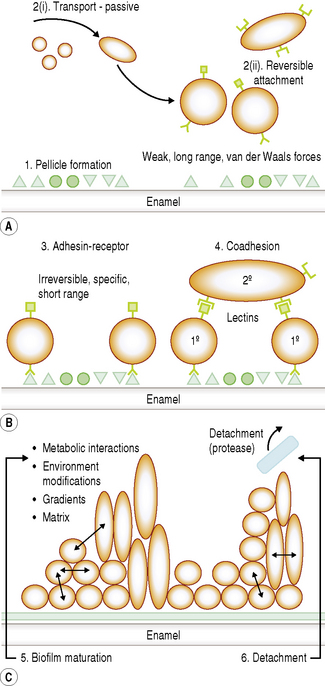
The development of a biofilm such as dental plaque can be subdivided arbitrarily into several stages. As a bacterium approaches a surface a number of specific and non-specific interactions will occur between the substratum and the cell, and these will determine whether successful attachment and colonization will take place. Distinct stages in plaque formation are summarized below, and are shown schematically in Fig. 5.2.
1 Acquired pellicle formation
Bacteria rarely come into contact with clean enamel. As soon as a tooth surface is cleaned, salivary proteins and glycoproteins are adsorbed forming a surface conditioning film which is termed the acquired enamel pellicle. The major constituents of pellicle are salivary glycoproteins, phosphoproteins and lipids, including statherin, amylase, proline-rich peptides (PRPs) and host defence components (Chs 2 and 4). Bacterial components such as glucosyltransferases (GTFs) and glucan have also been detected in pellicle, and play a significant role in attachment. Depending on the site, pellicle can also contain components from gingival crevicular fluid (GCF).
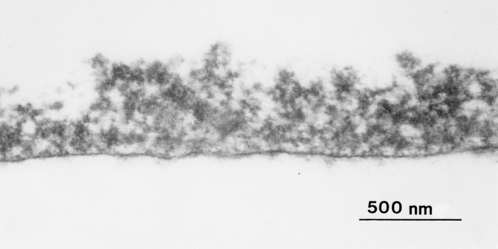
Pellicle formation starts within seconds of a clean surface being exposed to the oral environment. An equilibrium between adsorption and desorption of salivary molecules occurs after 90–120 minutes. The thickness of the pellicle is influenced by the shear forces at the site of formation. After 2 hours, the pellicle on lingual surfaces is 20–80 nm thick whereas buccal pellicles can be 200–700 nm deep. Depending on the site, further increases in depth can occur over time. Pellicle has an electron dense basal layer covered by a more loosely arranged globular surface (Fig. 5.3). When salivary molecules bind to the tooth surface, they can undergo conformational changes. This can lead to exposure of new receptors for bacterial attachment (cryptitopes; see later) or, in the case of glucosyltransferases, an altered activity resulting in the synthesis of a glucan with a modified structure. The molecular composition and physicochemical properties of pellicle are critical in determining the pattern of microbial colonization.
2 Transport of microorganisms and reversible attachment
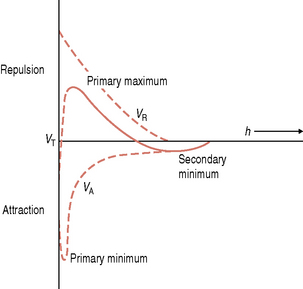
As the cell approaches the pellicle-coated surface, long range but relatively weak physicochemical forces are generated. Microorganisms are negatively-charged due to the molecules on their cell surface, while many proteins present in the acquired pellicle also have a net negative charge. The Derjaguin and Landau and the Verwey and Overbeek (DLVO) theory has been used to describe the interaction between an inert particle (as a microorganism might be envisaged at large separation distances) and a substratum. This theory states that the total interactive energy, VT, of two smooth particles is determined solely by the sum of the van der Waals attractive energy (VA) and the usually repulsive, electrostatic energy (VR). Particles in aqueous suspension and surfaces in contact with aqueous solutions can acquire a charge due to, for example, the preferential adsorption of ions from solution or the ionization of certain groups attached to the particle or surface. The charge on a surface in solution is always exactly balanced by an equivalent number of counter ions; the size of this electrical double layer is inversely proportional to the ionic strength of the environment. As a particle approaches a surface, therefore, it experiences a weak van der Waals attraction induced by the fluctuating dipoles within the molecules of the two approaching surfaces. This attraction increases as the particle moves closer to the substratum. A repulsive force is encountered if the surfaces continue to approach each other, due to the overlap of the electrical double layers. Curves can be plotted to show the variation of the total interactive energy, VT, of a particle and a surface with the separation distance, h (Fig. 5.4). A net attraction can occur at two values of h; these are referred to as the primary minimum (h very small) and the secondary minimum (h = 10–20 nm) and are separated by a repulsive maximum. The reversible nature of these initial interactions suggests that the primary minimum is not usually encountered while the high ionic strength of saliva increases the likelihood that oral bacteria could be retained near a surface by a secondary minimum area of attraction (e.g. about 10–20 nm from the surface).
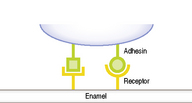
3 Pioneer microbial colonizers and irreversible attachment (adhesin–receptor interactions)
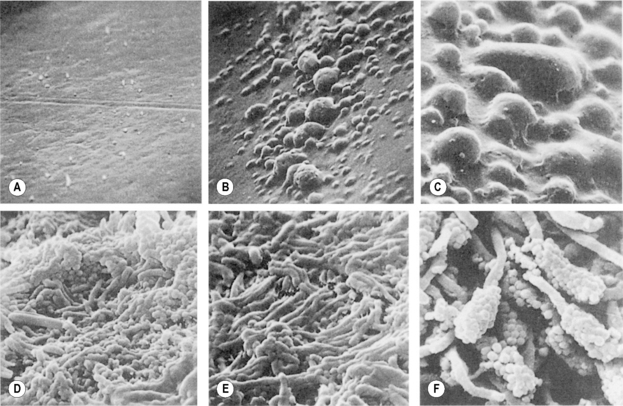
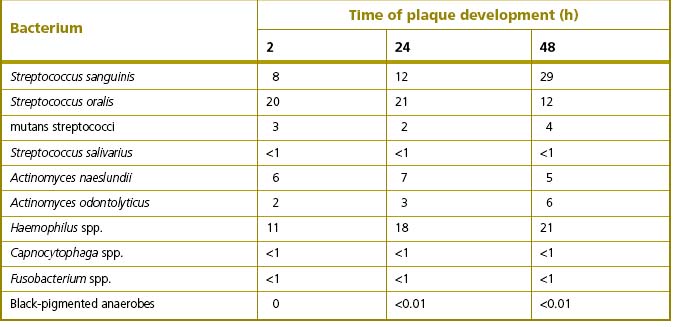
Irrespective of the type of tooth surface (enamel or cementum), the initial colonizers constitute a highly selected part of the oral microflora. Within minutes, coccal bacteria appear on the surface (Fig. 5.5A), and these pioneer organisms are mainly streptococci, especially members of the mitis-group of streptococci (e.g. S. sanguinis, S. oralis and S. mitis biovar 1) (Table 5.2). Streptococcus sanguinis and S. oralis produce an IgA1 protease which may help them to survive and overcome a key element of the host defences during the early stages of plaque formation. Actinomyces spp. are also commonly isolated after 2 hours, as are Haemophilus spp. and Neisseria spp., while obligately anaerobic species are detected only rarely at this stage and are usually in low numbers. Some aggregates of mixtures of cells may also attach.
Once attached, these pioneer populations start to divide and form microcolonies; these early colonizers become embedded in bacterial extracellular slimes and polysaccharides together with additional layers of adsorbed salivary proteins and glycoproteins (Fig. 5.5, B and C). The early streptoccocal colonizers (mitis-group of streptococci) possess a range of glycosidase activities that enable them to interact and use salivary glycoproteins as substrates (Ch. 4; see Fig. 5.13 later). The fastest rates of multiplication occur during these early stages of plaque formation; the doubling times of pure cultures of S. mutans and A. naeslundii were 1.4 h and 2.7 hours, respectively, in studies using rodents.
The irreversible attachment of cells to the tooth involves specific, short range, stereochemical interactions between components on the microbial cell surface (adhesins) and complementary receptors in the acquired pellicle. These specific interactions contribute to the often observed associations (tropisms; Ch. 4) of certain organisms with a particular surface or habitat.
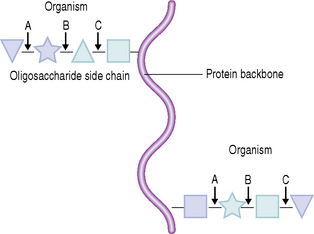
Several examples of these molecular interactions have been characterized, and some are listed in Table 5.3. Other examples of adhesins and their complementary receptors are described in Chapter 4. As examples, adhesins on S. gordonii can bind to α-amylase, while A. naeslundii and F. nucleatum interact with statherin. Streptococcus mutans (but not S. sobrinus), P. gingivalis, P. loescheii and P. melaninogenica adhere preferentially to hydroxyapatite coated with PRPs, although different regions of the molecule bind to particular organisms. Colonization by S. sobrinus is more dependent on sucrose-mediated mechanisms, including the interaction of glucans with receptors such as glucan binding proteins. Some adhesion mechanisms involve lectin-like bacterial proteins which interact with carbohydrates, or oligosaccharides, on glycoproteins adsorbed to the tooth surface. Thus, S. sanguinis can bind to terminal sialic acid residues in adsorbed salivary glycoproteins, while S. oralis expresses either a galactose-binding lectin or a lectin that interacts with a trisaccharide structure containing sialic acid, galactose and N-acetylgalactosamine.
Table 5.3 Some examples of host-bacterial interactions involved in adhesion.
| Bacterium | Adhesin | Receptor |
|---|---|---|
| Streptococcus spp. | antigen 1/11 | salivary agglutinin |
| Streptococcus spp. | LTA | blood group reactive glycoproteins |
| Mutans streptococci | glucan binding protein | glucan |
| Streptococcus parasanguinis | 35 kDa lipoprotein | fibrin, pellicle |
| Actinomyces naeslundii | type 1 fimbriae | proline-rich proteins |
| Porphyromonas gingivalis | 150 kDa protein | fibrinogen |
| Prevotella loescheii | 70 kDa lectin | galactose |
| Fusobacterium nucleatum | 42 kDa protein | coaggregation with Porphyromonas gingivalis |
A number of proteins in the cell walls of streptococci have been identified as adhesins. A high molecular weight protein from S. mutans, (termed antigen I/II, B, P1 or Pac), interacts with salivary agglutinins. Antibody directed against this surface molecule can block the adhesion of S. mutans to saliva-coated surfaces. The large size of some of the bacterial cell wall proteins means that they may be involved in more than one function. For example, a protein of S. gordonii can interact both with salivary proteins and with A. naeslundii (coaggregation). Some of the other adhesins found on S. sanguinis and S. gordonii are now known to be lipoproteins (Ch. 4). Streptococci can use multiple adhesins to bind to saliva-coated surfaces; as an example, S. sanguinis can adhere via lectin-like, hydrophobic and/or specific protein (adhesin) interactions.
A critical factor in plaque formation concerns the site at which the specific interactions between bacterial adhesins and host receptors take place. The host-derived receptors reside on molecules that are not only adsorbed to the tooth surface but which are also freely accessible in solution in saliva. Some of these molecules are designed to aggregate bacteria in solution, thereby facilitating their removal from the mouth by swallowing (see Ch. 2). For plaque formation to proceed, however, it is implicit that not all bacteria are aggregated in saliva before they reach the tooth surface. A novel mechanism may function to overcome this problem. It was found that although A. naeslundii could bind to the acidic PRPs when the latter were bound to a surface, cells did not interact with these proteins in solution. It has been proposed that hidden molecular segments of PRPs become exposed only when the proteins are adsorbed to hydroxyapatite, as a result of conformational changes. Such hidden receptors for bacterial adhesins have been termed ‘cryptitopes’. In this way, a selective mechanism for facilitating natural plaque formation has evolved by which the host can promote the attachment of specific bacteria without compromising this process in the planktonic phase. Adhesins which recognise cryptitopes in surface-associated molecules would provide a strong selective advantage for any microorganism which colonizes a mucosal or tooth surface. Another example of a cryptitope involving conformational change is the binding of members of the mitis-group of streptococci to fibronectin when complexed to collagen but not to fibronectin in solution. This might also be a mechanism enabling certain oral streptococci to colonize dam/>
Stay updated, free dental videos. Join our Telegram channel

VIDEdental - Online dental courses