Chapter 4 Acquisition, adherence, distribution and metabolism of the oral microflora
Acquisition of the resident oral microflora
Acquisition depends on the transmission of microorganisms to the site of potential colonization. Initially, in the mouth, this is by passive inoculation from the mother, from other individuals in close proximity to the baby, and from ingested milk and water. Acquisition of microorganisms such as yeasts and lactobacilli from the birth canal itself may be only transient, but the role of saliva in the process of acquisition has been confirmed conclusively. The ability to type strains (Fig. 3.1) has confirmed the transfer of Streptococcus salivarius, mutans streptococci and some other species from mother to child via saliva. Similarly, comparisons of the DNA fingerprints (genotyping) of a variety of oral bacterial species have shown that the same digest pattern (and hence presumably the same clonal type) is commonly found within family groups, and that different patterns are usually observed between such groups. Nevertheless, strains of some bacteria can be acquired occasionally by young children from other family members, while some strains seem to be distinct from any found in close relatives. The genotypes of mutans streptococci found in children were identical to those of their mothers in 71% of 34 infant-mother pairs examined (vertical transmission). Little evidence of father-infant (or father–mother) transmission of mutans streptococci was observed, although horizontal transmission between spouses, and vertical transmission within family units, can occur with some periodontal pathogens, such as Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans.
Pioneer community and microbial succession
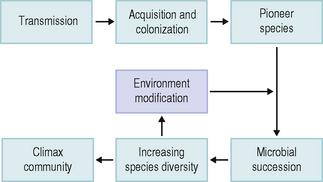
As outlined above, the pioneer community influences the pattern of microbial succession. This involves the progressive development of a pioneer community (containing few species) through several stages in which the number of microbial groups increases, until an equilibrium is reached; this is termed the climax community, and generally has a high species diversity (Fig. 4.1). Succession is associated with a change from a site possessing few niches (roles – Ch. 1) to one with a multitude of potential niches. A climax community reflects a highly dynamic situation between the host, the environment and the microflora, and must not be regarded as a static state.
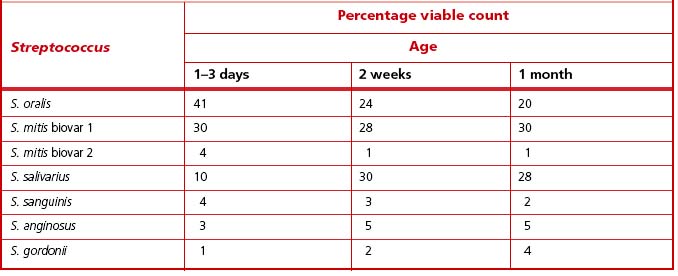
The oral cavity of the newborn contains only epithelial surfaces for colonization. The pioneer populations consist of mainly aerobic and facultatively anaerobic species. In full-term babies, a range of streptococcal species have been recovered during the first few days of life, and S. oralis, S. mitis biovar 1 and S. salivarius were numerically dominant (Table 4.1). The diversity of the streptococcal microflora increases with time; after one month, all babies were colonized by at least two species of Streptococcus, with S. salivarius and S. mitis biovar 1 being isolated most commonly (Table 4.1). In contrast, the prevalence of S. sanguinis increased only after 12 months. Actinomyces odontolyticus was isolated from oral mucosal surfaces in infants as young as two months; A naeslundii was generally only present in older babies (ca. 12 months old) once teeth had erupted.
The diversity of the pioneer oral community increases during the first few months of life, with some Gram negative obligate anaerobes appearing. Prevotella melaninogenica was the most frequently isolated anaerobe, being recovered from 76% of edentulous infants (mean age = 3 months; range: 1–7 months) (Table 4.2). Other commonly isolated bacteria were Fusobacterium nucleatum (present in 67% of infants), Veillonella spp. (63%), and non-pigmented Prevotella spp. (62%), while Eikenella corrodens and Wolinella succinogenes were found only in a single mouth (Table 4.2). The number of different anaerobes in the same mouth varied from 0–7 species.
Table 4.2 The effect of tooth eruption on the composition of the cultivatable oral microflora in 21 young children.
| Bacterium | Percentage isolation frequency | |
|---|---|---|
| Mean age | ||
| 3 months | 32 months | |
| Prevotella melaninogenica | 76 | 100 |
| non-pigmented Prevotella | 62 | 100 |
| Prevotella loescheii | 14 | 90 |
| Prevotella intermedia | 10 | 67 |
| Prevotella denticola | ND* | 71 |
| Fusobacterium nucleatum | 67 | 100 |
| Fusobacterium spp. | ND | 71 |
| Selenomonas spp. | ND | 43 |
| Capnocytophaga spp. | 19 | 100 |
| Leptotrichia spp. | 24 | 71 |
| Campylobacter spp. | 5 | 43 |
| Eikenella corrodens | 5 | 57 |
| Veillonella spp. | 63 | 63 |
The same infants were followed longitudinally during the eruption of the primary dentition. Gram negative obligately anaerobic bacteria were isolated more commonly, and a greater diversity of species were recovered from around the gingival margin of the newly erupted teeth (mean age of the infants = 32 months) (Table 4.2). These findings confirm that the eruption of teeth has a significant ecological impact on the oral environment and its resident microflora.
Allogenic and autogenic microbial succession
The establishment of a climax community at any oral site involves a series of phases of development during which the complexity of the microflora increases (microbial succession; Fig. 4.1). Two distinct types of succession have been identified (Fig. 4.2). In allogenic succession, factors of non-microbial origin are responsible for an altered pattern of community development. For example, the frequency of detection of mutans streptococci and S. sanguinis in the mouth increases markedly once hard, non-shedding surfaces are present. This is usually following tooth eruption, but it can also occur after the insertion of dentures or removable orthodontic appliances, and acrylic obturators in children with cleft palate.
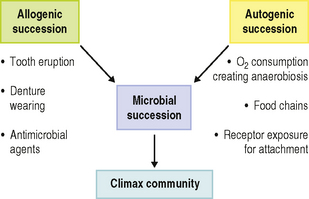
Fig. 4.2 Role of autogenic and allogenic succession in the development of oral microbial communities.
The increase in number and diversity of obligate anaerobes once teeth are present is an example of autogenic succession in which community development is influenced by microbial factors (Fig. 4.2). The metabolism of the aerobic and facultatively anaerobic pioneer species lowers the redox potential in plaque and creates conditions suitable for colonization by strict anaerobes (Ch. 5). Other examples of autogenic succession are the development of food chains and food webs, whereby the metabolic end product of one organism becomes a primary nutrient source for another:
A further example of autogenic succession is the exposure of new receptors for bacterial adhesion (‘cryptitopes’; Ch. 5) following microbial modification of host macromolecules.
The gross composition of the oral microflora can remain relatively stable over time at individual sites, especially when analyzed at the genus or species level. Ribotyping (Ch. 3) can discriminate among strains within a species on the basis of genetic variation, thereby allowing specific clonal types to be recognised. Relatively few clones are found within species of pathogenic bacteria, and a limited number of these may be responsible for the majority of infections. In contrast, species that comprise the resident human microflora generally display large numbers of clones, and this may be a strategy to help such species evade the host defences. Clones of some species appear to persist for long periods at a site whereas others appear to be transient, and undergo replacement by fresh clones. For example, clonal replacement appears to maintain S. mitis biovar 1 in the mouth of neonates; 93 clonal types were detected among 101 strains of S. mitis colonizing 40 infants over a one month period. The clonal types that could be isolated were found to vary at different sampling times, suggesting that individual clones did not persist and were replaced by new clones. In a further study of the clonal diversity of S. mitis biovar 1, limited sharing of genotypes was found among three members of a particular family, and each individual carried between 6 and 13 types. Differences were also found between isolates recovered from the pharyngeal and buccal mucosa of the same individual. The reasons for, and the mechanisms involved in, the persistence of certain clones of some species but the continual turnover of clones of other species is not yet understood. It may be that the niche (function) occupied by these clonal types is similar, but that turnover may represent a longer-term survival strategy. Wide variations in the expression of carbohydrate and protein antigens have been found among the different genotypes of S. mitis biovar 1 from the family group, suggesting that this ‘turnover’ might play a role as an ‘immune-evasion’ mechanism for this species.
In contrast to the situation for S. mitis, a particular clone of Aggregatibacter actinomycetemcomitans has been implicated in an aggressive form of periodontitis in young adults (Ch. 6). The JP2 clone has a 530 base pair deletion in the promoter region of the leukotoxin gene operon, resulting in increased production of the leukotoxin. Adolescents harbouring this clone are more likely to suffer loss of attachment between the tooth and the periodontium than those who have non-JP2 types. In the future, the ability to recognise virulent clones of an organism may result in the better diagnosis and prediction of sites/individuals at risk of disease.
Ageing and the oral microflora
In adults, the composition and proportions of the resident oral microflora remain reasonably stable over time and this microflora coexists in relative harmony with the host. This stability (termed microbial homeostasis; Ch. 5, Fig. 5.18) is not a passive response to the environment, but is due to a dynamic balance being achieved from numerous inter-bacterial and host- bacterial interactions. The diversity of the oral microflora in a healthy individual is typically between 50–100 species.
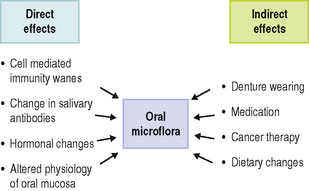
Some variations in the oral microflora have been discerned in later life and can be attributed to both direct and indirect effects of ageing (Fig. 4.3). In the case of the latter, variations can occur if the habitat or environment is severely perturbed. For example, the risk of cancer rises with age, and cytotoxic therapy or myelosuppression combined with the disease itself is associated with the increased carriage of Candida albicans and non-oral opportunistic pathogens such as enterobacteria (e.g. Klebsiella spp., Escherichia coli, Pseudomonas aeruginosa) and Staphylococcus aureus. The wearing of dentures also increases with age and this also promotes colonization by C. albicans. Many elderly subjects take a variety of medications, the side-effects of which can reduce the flow of saliva and thereby perturb the normal balance of the resident oral microflora.
Methods of determining the composition of the resident oral microflora
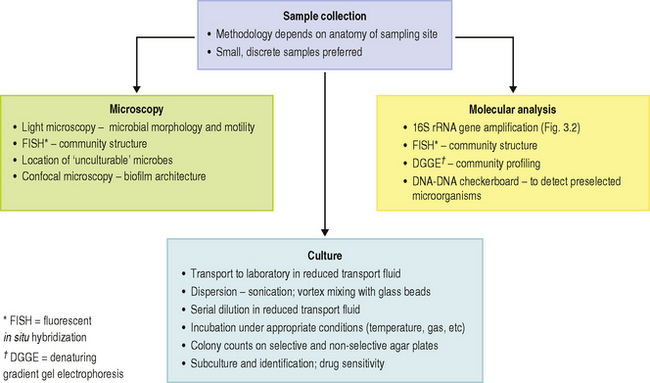
There are a number of challenges when attempting to determine the composition of the microbial communities at sites in the mouth. These range from the basic problem of removing the majority of the microorganisms from their habitat (many of which are by necessity bound tenaciously to a surface or to each other, and the site may be difficult to access) to their eventual identification (Table 4.3; Ch. 3). The main approaches to determining the microbial composition of the oral microflora are illustrated in Fig. 4.4, while the main differences between culture-dependent and -independent approaches were highlighted in Figure 3.2. Some of the issues are discussed in more detail below:
Table 4.3 Some properties of the oral microflora that contribute to the difficulty in determining its composition.
| Property | Comment |
|---|---|
| High species diversity | The oral microflora, and especially dental plaque, consists of a diverse number of microbial species, some of which are present only in low numbers. |
| Surface attachment/coaggregation (coadhesion) | Oral microorganisms attach firmly to surfaces and to each other and, therefore, have to be dispersed without loss of viability. |
| Obligate anaerobes | Many oral bacteria lose their viability if exposed to air for prolonged periods. |
| Fastidious nutrition/unculturable | Some bacteria are difficult to grow in pure culture and may require specific cofactors etc. for growth. Some groups (e.g. certain spirochaetes; TM7 group) cannot as yet be cultured in the laboratory. |
| Slow growth | The slow growth of some organisms makes enumeration time consuming, (e.g. they may require14–21 days incubation). |
| Identification | The classification of many oral microorganisms still remains unresolved or confused; simple criteria for identification are not always available (particularly for some obligate anaerobes). |
Sample taking
The microflora can vary in composition over relatively small distances. Therefore, large plaque samples or a number of smaller pooled samples from different sites can be of little value because important site-specific differences will be obscured. Consequently, small samples from discrete sites are preferable, but the method of sampling will depend on the anatomy and properties of the site to be studied.
The oral mucosa can be sampled by swabbing, direct impression techniques, or by removing epithelial cells by scraping or scrubbing with a blunt instrument into a container. Microbial counts can then be related to a fixed area or to an individual epithelial cell. Saliva can be collected by expectoration into a sterile container; the saliva flow can be at a normal resting rate (unstimulated) or it can be stimulated by chemical means or by chewing. Although a greater volume is collected by stimulation, such samples will also contain many more organisms that have been dislodged from oral surfaces.
There is no universally accepted way of sampling dental plaque. The accessible smooth surfaces of enamel pose few problems and a range of dental instruments have been used. Dental probes, scalers, dental floss and abrasive strips have been used to remove plaque from approximal surfaces between teeth. Fine probes, pieces of wire, blunt hypodermic needles, and toothpicks have been used to sample plaque from fissures, although the amount of biofilm removed can depend on the anatomy of the site. Subgingival plaque is difficult to sample because of the inaccessibility and anaerobic nature of the site. High numbers of obligately anaerobic bacteria are found in the gingival crevice and periodontal pocket, most of which will lose their viability if exposed to air. In disease, the anatomy of the site means that those organisms at the base of the pocket, near the advancing front of the lesion, are likely to be of most interest (Ch. 6). Again, it is important to avoid removing plaque from other areas within the pocket so as not to obscure significant relationships between particular bacteria and disease. A common approach is to insert paper points into pockets but the number of firmly adherent organisms removed from the root of the tooth will be small. Samples have also been taken by irrigation of the site and retrieval of the material through syringe needles; however, this method will obviously remove plaque from the whole depth of the pocket. A particularly sophisticated method employed a broach kept withdrawn in a cannula that was flushed constantly with oxygen-free nitrogen. The broach was used to sample plaque only when the cannula was in position near the base of the pocket. After sampling, the broach was retracted into the cannula and withdrawn. Another approach has been to use a curette or scaler after the supragingival area has been cleared. The scaler tips can be detached and placed immediately in gas-flushed tubes containing reduced (anaerobic) transport fluid for delivery to the laboratory. Alternatively, when periodontal surgery is needed, plaque has been removed from extracted teeth or from surfaces exposed when ‘gingival flaps’ are reflected. It is important to appreciate, particularly when comparing studies in which different sampling procedures have been used, that the results will, to a certain extent, reflect the method adopted.
Cultivation
Once dispersed, samples are usually serially diluted in a suitable fluid (usually a transport fluid) and aliquots are spread on to a number of freshly prepared, pre-reduced agar plates, and incubated to allow cells to form microbial colonies. These media are designed to grow either (a) the maximum number of bacteria (e.g. blood agar) or, (b) only a limited number of species (selective media) in order to recover minor components of the microflora. For example, the addition of vancomycin to blood agar plates will inhibit most Gram positive bacteria, while a high sucrose concentration encourages the growth of oral streptococci, and plates with a low pH favour lactobacilli. It should be emphasised that these media are selective and not specific for any type of microbe. The identity of the colonies on these plates must be confirmed; colonial appearance or mere growth on a particular medium is not diagnostic. Media need to be incubated for different times and under different atmospheric conditions depending on the bacteria being cultivated. For example, 7–14 days incubation at 37 °C in an anaerobic jar or cabinet filled with a gas mix containing CO2/H2/N2 will be needed to grow some obligate anaerobes; in contrast, Neisseria require only 2 days incubation in air. Some organisms are capnophilic (CO2-loving) and grow optimally in 10% CO2 in air. Other media require supplementation with growth factors in order to enable certain fastidious organisms to grow (Ch. 3). As mentioned previously, at present only about 50% of the resident oral microflora can be cultured in the laboratory.
Enumeration and Identification
The first level of discrimination involves the Gram staining of subcultured colonies; bacteria are then grouped according to whether their cells are Gram positive or Gram negative, and are rod- or coccal-shaped. This dictates which tests will be necessary to achieve speciation. Some bacteria can be identified using simple criteria, for example, sugar fermentation tests or the detection of preformed enzymes using commercial kits. Other bacteria require a more sophisticated approach such as the application of gas–liquid chromatography to determine their acid end-products of metabolism (Table 3.2). The use of probes (antibody or oligonucleotide) on isolates can speed up identification and distinguish between strains of the same species (for example, by ribotyping); this level of discrimination is essential for studies of bacterial transmission. Other issues relating to bacterial identification are discussed in Chapter 3.
Microscopy
As an alternative to many of the lengthy steps associated with conventional culture techniques, the principal morphological groups of bacteria can be determined using light microscopy (Fig. 4.4). Dark-field illumination or phase contrast techniques have been used to quantify the numbers of motile bacteria (including spirochaetes) directly in dental plaque (particularly from subgingival sites). Such organisms are related to the severity of some periodontal diseases, and this approach has been used in the clinic to monitor sites undergoing treatment. However, most of the putative pathogens cannot be recognized by morphology alone. To overcome this problem, cells can be identified by reaction with antisera (monoclonal or specific polyclonal), oligonucleotide probes or tagged with a fluorescent label (Fig. 3.3).
Scanning and transmission electron microscopy have proved useful in studying plaque formation, and have also been used to show that bacteria invade gingival tissues in aggressive forms of periodontal disease. Electron dense markers (ferritin, peroxidase, gold granules) conjugated with antibodies can label specific surface antigens on bacteria and facilitate their identification within plaque. Electron microscopy requires samples to be processed before viewing which can distort the structure of plaque. Non-invasive techniques such as confocal laser scanning microscopy are now being used, with and without the use of specific probes (antibody or oligonucleotide), to determine the true architecture of plaque and the location of selected bacteria within the biofilm (Ch. 5). Confocal microscopy involves the generation of numerous focussed images throughout the depth of an untreated specimen (optical sections); image analysis software is then used to combine these sections and reconstruct the three-dimensional structure of the original specimen (for an example, see Fig. 4.6). Confocal microscopy has shown that plaque may have a more open architecture than previously thought from studies involving electron microscopy.
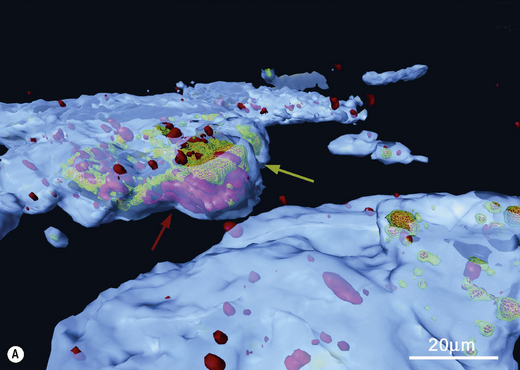
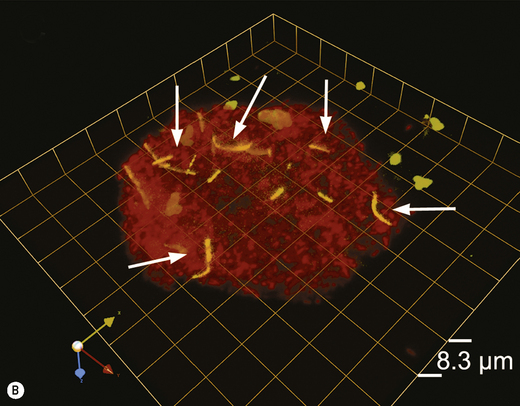
Fig. 4.6 Intracellular colonization of buccal epithelial cells. (A) Three-dimensional reconstruction of a buccal epithelial cell (BEC). Bacteria recognized only by a universal bacterial probe are shown in solid red, while co-localization of the Aggregatibacter actinomycetemcomitans and universal probes is depicted by a green wireframe over a red interior. Reconstructed BEC surfaces are presented in blue. The red and green colours are muted when bacterial masses are intracellular, and brighter when bacteria appear to project out of the surface. The />
Stay updated, free dental videos. Join our Telegram channel

VIDEdental - Online dental courses