Fig. 1.1
Initial stages of biofilm formation. Schematic outlining the general approaches of initial cellular interaction of planktonic cells with coated substrates. In the initial phase, a “clean” surface is coated with environmental elements. At the second stage, a planktonic cell that approaches the coated surface initiates adhesion by adjusting a number of regulatory mechanisms known as surface sensing. In the following stage, irreversible adhesion occurs by association of specific cell components such as pili, flagella, exopolymers, etc. Lastly, co-adhesion with other organisms is achieved by specific interspecies interactive mechanisms

Fig. 1.2
Biofilm growth and maturation. Image sections showing reconstructed three-dimensional biofilm images at a magnification of ×100. Biofilms were stained with LIVE/DEAD stain, resulting in live and dead bacteria appearing as green or red, respectively. 3D images show confocal images of biofilm formation by oral bacteria at 1, 3, 5, and 7 days of growth, respectively. Upper image shows the first stage of biofilm growth at day 1; second and third images show subsequent stages of biofilm formation at day 3 and 5, respectively. Bottom image shows the fourth stage of biofilm formation at day 7. Damaged organisms appear red and undamaged organisms appear green
Biofilms initiate formation when a free-floating cell (cell in planktonic state) is deposited on a substratum coated with an organic conditioning polymeric matrix or “conditioning film” (Fig. 1.1). Conditioning films are composed by constituents of the local environment like water, salt ions, albumin, or fibronectin. When the first bacterial cells arrive, there is a weak and reversible contact between the cell and the conditioning film resulting from physical interactions such as Brownian motion, gravitation, diffusion, or electrostatic interactions [21]. Specific interactions with bacterial surface structures such as flagella and pilus are also important in the initial formation of a biofilm. The next step is when the adhesion of the cell to the substrate becomes irreversible. This is partly due to surface appendages overcoming the repulsive forces between the two surfaces and also helped by the sticky exopolymers secreted by the cells. These hydrophilic exopolymers have a complex and dynamic structure [22].
As depicted in Fig. 1.2, the second part of the formation of a biofilm comprises its growth and development. Development of a biofilm occurs as a result of adherent cells replicating and by additional cells adhering to the biofilm [37]. This is an overall dynamic process where many microorganisms co-adhere to one another and interact in the now active communities. Consequently during growth some cells will be detaching from the biofilm over time [6, 8, 28, 47].
Biofilms Developed in Root Canals
As surface-associated microbial communities are the main form of colonization and retention by oral bacteria in the mouth, it is not unreasonable to assume that biofilms also form in root canals having the same properties as the parent communities colonizing the enamel and cementum surfaces [10]. Microorganisms have been found to colonize by adhering to dentine walls in all the extension of the root canals. These aggregations of microorganisms have been observed adhered to the inner walls of complex apex anatomies and accessory canals [61, 71]. When these biofilm communities are formed on surfaces located beyond the reach of mechanical removal and the effects of antimicrobials, host-derived proteins from remaining necrotic tissues and bacterially produced adhesive substances will provide the proper prerequisites for the survival of microbes.
In 2004, Svensäter and Bergenholtz [83] proposed a hypothesis for biofilm formation in root canals. Biofilm formation in root canals is probably initiated just after the first invasion of the pulp chamber by oral organisms following the pulp tissue inflammatory breakdown. The inflammatory lesion frontage will then move successively towards the apex providing the fluid vehicle for the invading organisms so these can multiply and continue attaching to the root canal walls. Interestingly, bacteria have been observed to detach from inner root canal surfaces and occasionally mass in the inflammatory lesion per se [61, 71]. This observation could explain how the inflammatory lesion front serves as a fluid source for bacterial biofilm detachment and colonization of other remote sites in the root canal.
Resistance to Antimicrobials
Biofilm bacteria usually have an increased resistance to antimicrobial agents, in some cases up to 1,000-fold greater than that of the same microorganisms living in liquid suspension [27, 38]. Biofilms formed by oral bacteria are more resistant to chlorhexidine, amine fluoride, amoxicillin, doxycycline, and metronidazole than planktonic cells [46, 75]. Therefore, it is reasonable to assume that biofilms formed in root canals will also share the same resistant properties as oral bacteria, a fact that will affect the overall prognosis of root canal treatments. The high resistance capacity of biofilm communities from root canal bacteria was shown in a series of experiments that tested the resistance of biofilms formed by bacteria isolated from infected root canals to alkaline stress [12]. In this study, the viability of susceptible root canal strains in planktonic cultures was found to be considerably increased when the same strains were exposed to the same alkaline stress in biofilms.
The reasons for the increased resistance of bacteria when forming a biofilm are believed to be multiple, and currently, there is no generally agreed upon specific mechanism(s). It would seem that resistance is dependent in multiple factors such as the substrate, microenvironment, and age of the biofilm [80, 81]. There are, however, a number of known mechanisms that account for this broad resistance and can be divided in two main groups: (a) physical and (b) acquired. The physical protection is mainly related to the impaired penetration of antibiotics through the biofilm matrix. As it is illustrated in Fig. 1.3, acquired resistance is divided into three subcategories: differentiation of cells with low metabolic activity, differentiation of cells that actively respond to stress, and differentiation of cells with a very high persistent phenotype.
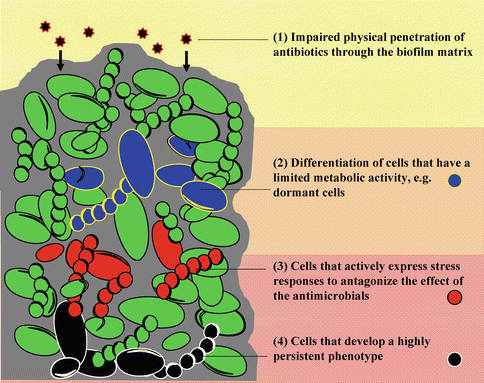

Fig. 1.3
Mechanisms of resistance by biofilm bacteria. The illustration depicts different mechanisms of resistance by biofilm bacteria. Slow or incomplete penetration of antimicrobials through the matrix (1). Concentration gradients of metabolites and waste will form zones where subpopulations of bacteria are differentiated. These subpopulations have different antimicrobial resistance capacities depending on their metabolic activity (dormant cells labeled blue) (2) or if they develop an active stress response mechanism (red cells) (3). Finally, a subpopulation of persister cells may also develop (black cells) (4)
Physical Barrier to the Penetration of Antimicrobials in Biofilms
The main barrier that will hinder the penetration of antibiotics into the biofilm is the extracellular matrix [7, 26]. The extracellular matrix is the backbone of the biofilm and it is very complex in its composition, wide ranging between polysaccharides, proteins, nucleic acids, and lipids. The extracellular polymeric substances (EPS) provide not only physical and adhesive stability to the biofilm, but they also form the scaffold for the three-dimensional architecture that interconnects and organizes cells in biofilms [26].
Critical to matrix function is the distribution of the varied molecular-complex components that influences the developmental, homeostatic, and defensive processes in biofilms. Because of the marked diversity of EPS – inclusive of glycoproteins, proteoglycans, and insoluble hydrophobic polymers, among other components depending on the species involved – it is not surprising that this slimy substance delays considerably the diffusion of antimicrobials [81]. For example, it has been directly observed a profound retardation in the delivery of a penicillin antibiotic from penetrating a biofilm formed by a betalactamase-positive bacterium [3].
Due to the physical protection provided by the biofilm matrix, intense research is ongoing that aim to target the identification of novel matrix components. This novel research on matrix components will provide evidence for the identification and application of matrix-degrading enzymes that may prevent formation and/or activate dispersal of biofilms [45]. Some examples of novel biofilm matrix components that are currently studied are listed in Table 1.1.
Table 1.1
Novel biofilm matrix components recently found and under current research
|
Biofilm matrix component
|
Biofilm-forming species
|
Reference
|
|---|---|---|
|
Exopolysaccharide
|
Bacillus subtilis (NCIB3610)
|
[7]
|
|
Poly-gamma-DL-glutamic acid
|
B. subtilis (RO-FF-1)
|
[79]
|
|
Poly-N-acetyl glucosamine (PNAG)
|
S. aureus
|
[66]
|
|
Amyloid fibers of the protein TasA
|
B. subtilis
|
[72]
|
|
Protein BapL
|
L. monocytogenes
|
[39]
|
|
BAP proteins
|
S. aureus
|
[87]
|
|
Extracellular protein, MabA
|
Lactobacillus rhamnosus
|
[88]
|
|
Extracellular DNA (eDNA)
|
Bacillus cereus, S. aureus, and L. monocytogenes
|
State of Nutrient Deprivation and Dormancy
It has been observed that throughout the various sections of the biofilm, cells are in different physiological states. Cells at the base of the film, for example, may be dead or lysing, while those near the surface may be actively growing [19, 80]. However, the majority of time cells in biofilms are in a dormant state that is equivalent to cells in the stationary phase of growth [64, 65]. In particular this dormant state is hypothesized to be common in biofilms that are formed in microenvironments where nutrients are scarce, such as treated root canals of teeth [14]. This dormant physiological state related to the general stress response and associated survival responses may offer an explanation for the resistance of biofilm cells to antimicrobials.
Bacteria under the stress of nutrient deprivation have developed efficient adaptive regulatory mechanisms to modify their metabolic balance away from biosynthesis and reproduction [40, 73]. One such mechanism involves the stringent response, a global bacterial response to nutritional stress that is mediated by the accumulation of the alarmones guanosine tetraphosphate and guanosine pentaphosphate, collectively known as (p)ppGpp [25, 68, 85]. For example, (p)ppGpp plays an important role for low-nutrient survival of E. faecalis, an organism that is known to withstand prolonged periods of starvation and remain viable in root-filled teeth for at least 12 months [58, 67]. Furthermore, the alarmone system (p)ppGpp has also a profound effect on the ability of E. faecalis to form, develop, and maintain stable biofilms [15]. These improved understanding of the alarmone mechanisms underlying biofilm formation and survival by E. faecalis may facilitate the identification of pathways that could be targeted to control persistent infections by this organism.
From the perspective of the persisting root canal flora, it is reasonable to assume that such dormant cells might “wake up” at some point in time and resume their metabolic activity to provoke periapical inflammation. Thus, from the metabolic perspective, the reactivation of dormant cells will render biofilm bacteria able to contribute to the persistence of inflammation. For example, a recent case report of a tooth that was adequately treated and showed no signs of disease revealed recurrent disease after 12 years. Histopathologic and histobacteriologic analyses showed a heavy dentinal tubule infection surrounding the area of a lateral canal providing evidence on the persistence of an intraradicular infection caused by bacteria possibly located in dentinal tubules [90].
The above hypothesis on the reactivation of biofilm cells was tested in a recent study [14]. Biofilm cultures of oral isolates of Streptococcus anginosus and Lactobacillus salivarius were forced to enter a state of dormancy by exposing them to nutrient deprivation for 24 h in buffer. After the starvation period the number of metabolically active cells decreased dramatically to zero and their cell membrane integrity was kept intact. Biofilm cells were then exposed to a “reactivation period” with fresh nutrients, but even after 96 h, the cultures were dominated by undamaged cells that were metabolically inactive. This phenomenon was not observed for cells in a planktonic state that were rapidly reactivated after 2 h. The data produced by this study showed that biofilm cells exhibit a slow physiological response and, unlike cells in planktonic culture, do not reactivate in short time periods even under optimal conditions. This observation highlights the difference in physiology between the biofilm and planktonic cultures and also confirms the slower physiological response of biofilm cells [53, 54], a mechanism that may account as a strategy of biofilm bacteria to resist stressful conditions.
Formation of Phenotypically Different Subpopulations
Bacteria within biofilms differ in their phenotype, depending on the spatial location of the cells within the community [81, 96]. There is now consistent evidence that has proven the presence of subpopulations of cells within biofilms that significantly differ in their antibiotic susceptibility [32, 41]. This phenomenon is correlated with differences in chemical concentration gradients that create unique microenvironments within biofilm communities. Simultaneously, adaptive variability allows the cells to respond to their local environmental conditions [69, 97]. Numerous studies have investigated the creation of these phenotypically different subpopulations and their mechanisms including genetic alterations, mutations, genetic recombination, and stochastic gene expression. For example, Weiser et al. described two distinct phenotypic variants in S. pneumoniae that switched between a phenotype with the ability to adhere and coexist among eukaryotic cells and a phenotype that was less capable to adhere but was better adapted to evade the host immune response during inflammation or invasive infection [94]. Of interest is the fact that both phenotypes of S. pneumoniae differed in their production of capsular polysaccharide having the inflammation-resistant phenotype an increased production of up to two to six times more capsular polysaccharide. These differences were accentuated by changes in the environmental concentration of oxygen; decreased oxygen levels correlated with an increase in capsular polysaccharide expression.
Interestingly, the formation of subpopulation in biofilms, where physiological differences are in play, has been demonstrated to occur in multispecies biofilms by root canal bacteria [11]. This was shown using four root canal bacterial isolates that, when cocultured, reacted concurrently to the absence of glucose in the culture medium. Although the overall cell viability of the four-species community was not affected by the lack of glucose, there was a significant variation in the 3D structure of the biofilms. In addition, patterns of physiologic adaptation by members of the community to the glucose-deprived medium were observed. The metabolic activity was concentrated in the upper levels of the biofilms, while at lower levels the metabolism of cells was considerably decreased. Subpopulations of species with high glycolytic demands, streptococcus, and lactobacilli were found predominating in the upper levels of the biofilms. This distinct spatial organization in biofilms grown in the lack of glucose shows a clear reorganization of the community in order to satisfy their members’ metabolic pathways in order to enable the long-term persistence of the community. This result lends support to the hypothesis that the reorganization of subpopulations of cells in multispecies biofilms is also important for survival to stress factors from the environment [76].
Bacterial Cells That Persist
Groups of cells have been found to persist following exposure to lethal doses of antibiotics and new growing populations appear in the culture [48, 49]. These persister cells (a) may represent cells in some protected part of their cell cycle, (b) are capable of rapid adaptation, (c) are in a dormant state, or (d) are unable to initiate programmed cell death in response to the stimulus [49]. Thus, such persister cells represent a recalcitrant subpopulation that will not die and are capable of initiating a new population with normal susceptibility once the antibacterial effect has been dissipated. To date, these cells have only been reported to occur after the exposure of a bacterial population to high doses of a single antimicrobial agent, which triggered the appearance of persister cells exhibiting multiple drug resistance [51]. The frequency of persister occurrence and the mechanism(s) involved in their appearance are unclear, although one hypothesis with Escherichia coli suggests that persister cells are regulated by the expression of chromosomal toxin–antitoxin genes [42]. In this case, the operon HipA seems to be responsible for tolerance to ciprofloxacin and mitomycin C in stationary-phase planktonic cells and E. coli biofilms [42]. It has also been proposed that the expression of toxins drives bacteria reversibly into the slow-growing, multiple drug-tolerant phenotypes by “shutting down” antibiotic targets [50]. In the context of root canal bacteria, the formation of such persisting populations that are capable of surviving imposed endodontic treatment measures, as rise of the alkaline levels due to application of calcium hydroxide [12], would explain how organisms are able to survive and remain in the environment until the effects of noxious stimuli have dissipated.
Methods to Study Bacteria in Biofilms
The previous discussion relative to the capacity of biofilm bacteria to resist exposure to antimicrobials indicates the importance of studying the physiological state of bacteria with respect to their potential level of activity in the disease processes. However, the exact description of the status of a microorganism can be complex especially in chronic infections such as apical periodontitis. Currently, a variety of microscopic in situ methods have been developed to identify subpopulations and assess the physiological status of bacterial cells in biofilms. Some of these methods include molecular markers to study cell membrane integrity, metabolic activity, or the identification of stress encoding genes.
SEM and LSM
Electron microscopy (EM) in the transmission and scanning mode allows higher magnifications of fixed and dehydrated samples and, in combination with specific detectors, analysis of the elemental composition in specific regions of the sample [92]. EM provides resolution and magnification to offer a more detailed insight into the ultrastructure of the biofilm as well as its environment (Fig. 1.4). One of the main drawbacks of this technique, however, is that it requires the sample to be dehydrated prior to its analysis.
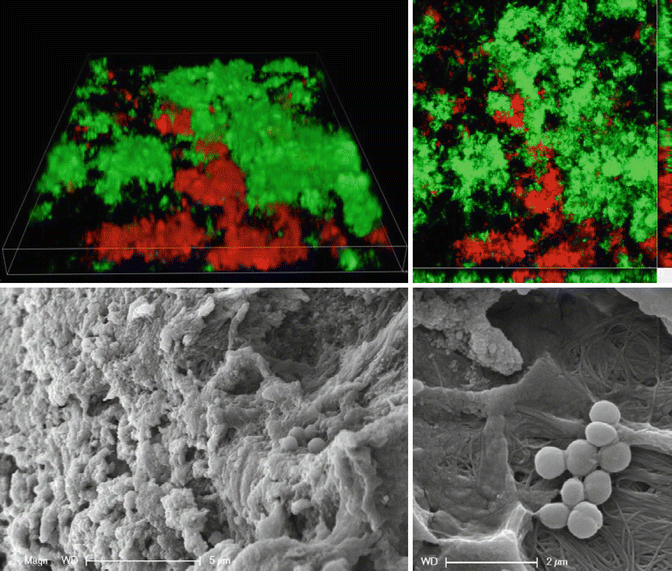

Fig. 1.4
LSM and SEM techniques. Observation of biofilm features by laser scanning microscopy and SEM. The panel above shows 3D reconstruction of biofilm structures labeled with LIVE/DEAD, a fluorescent marker of cell viability; green represents cells with intact cell membranes, while red represents cells with damaged membranes. The panel below shows ultrastructure of biofilms formed on apex of teeth as imaged by SEM. Scale bars: 5 and 2 μm (SEM images are courtesy of Dr. David Jaramillo)
The invention of laser scanning microscopy (LSM) in the 1980s caused a revolution in light microscopy. The LSM technique, usually called confocal laser scanning microscopy (CLSM), is nowadays the most important and indispensable tool for three-dimensional in situ imaging of microbial communities [9]. The LSM technique is mainly used to visualize multiple features in different channels that are spectrally resolved. By means of this imaging procedure, it is possible to analyze the structure, composition, microhabitats, activity, and processes using a variety of specific color probes. Finally, LSM allows the volumetric and structural quantification of multichannel signals in four dimensions [63]. One of the main disadvantages of LSM, however, is that the information captured from detailed ultrastructure of the biofilm is difficult. Very recently, this problem of LSM has been overcome with the advent of super-resolution microscopy (SRM). SRM encompasses a suite of cutting-edge microscopy methods able to surpass the resolution limits of common light microscopy [60]. It is foreseen that the application of SRM in combination with rRNA FISH (see below) would allow the tracking of ribosome-associated changes in activity levels and subcellular localization at the single-cell level [2].
rRNA Fluorescence In Situ Hybridization (FISH)
The combination of FISH with confocal laser scanning microscopy is one of the most powerful tools in modern microbiology as it allows visualization of specific subpopulation of cells while maintaining unaltered the 3D structure of the biofilm [1]. This high-throughput microscopy technique allows the specific detection and enumeration of biofilm subpopulations in situ in their natural environment without the need for cultivation [1]. Up to date a number of studies have demonstrated the direct use of CLSM-FISH on biofilm cultures growing in different surfaces [11, 23]. The most frequent application of FISH is the hybridization of oligonucleotide probes to ribosomal RNA, most often 16S but also 23S rRNA, for identification of single cells in their natural habitat [2]. Since ribosomes are the protein factories of all cells, their numbers are good proxies of general metabolic activity and of the physiological state of cells. Sequences of oligonucleotide probes targeting 16S rRNA have been developed for specific detection of different bacterial species and can be found in online databanks. In endodontics, FISH has been used to visualize and identify bacteria from periapical lesions of asymptomatic root-filled teeth [82]. Furthermore, biofilm models using CLSM-FISH can be of great advantage to investigate distribution of species in multispecies biofilms.
Markers of Cell Viability
Viability of bacteria is conventionally defined as the capacity of cells to perform all cell functions necessary for survival under given conditions [62]. The common method to assess bacterial viability is growth on plates, where the number of viable cells approximates the number of colony-forming units. In root canal infections, culture techniques have been the standard method used to assess bacterial viability. Once the living bacterial cells from root canals were isolated after growth on specific substrate, the metabolic properties of these bacterial isolates were then used to infer the potential roles of these and related microorganisms in a clinical context. Under some circumstances, however, such methods may underrepresent the number of viable bacteria for a variety of reasons, such as cases where slightly damaged organisms are present [4], the laboratory growth media employed are deficient for one or more essential nutrients required for the growth of some bacteria in the sample [93], or viable cells are present that have lost their ability to form colonies [95]. Furthermore, if the bacteria exist in a biofilm, they may assume a status of low metabolic activity similar to stationary-phase planktonic growth for the majority of time [65]. The bacteria in such low active states may be undetectable by regular culture techniques. The extent of this problem is reflected in the indiscriminate use of terms that are used to assess nonviable states, such as dead, moribund, starved, dormant, resting, quiescent, viable but not culturable, injured, sublethally damaged, inhibited, and resuscitable [62]. Many of these terms are used conceptually and do not reflect the actual knowledge of the exact viability state of the organism in question.
A number of viability indicators that can be assessed at the single-cell level without culturing cells have gained increased popularity in the latest years. These indicators are based mostly on fluorescent molecules, which can be detected with epifluorescence microscopy or laser scanning microscopy.
The LIVE/DEAD kit tests the integrity of the cell membrane by applying two nucleic acid stains, SYTO-9 and propidium iodide (PI), which can simultaneously detect dead/injured (fluorescent red by stain with PI) and intact cells (fluorescent green by staining with SYTO-9) [5]. This fluorescent probe has been used to assess the viability of root canal strains ex vivo [10] and to determine the autoaggregation and coaggregation of bacteria isolated from teeth with acute endodontic infections [44].
Alternative fluorescent probes to test bacterial viability are those that target specific cell metabolic functions, such as the tetrazolium salts 2-(4-iodophenyl)-3-(4-nitrophenyl)-5-phenyl tetrazolium chloride (INT) and 5-cyano-2,3-ditolyl tetrazolium chloride (CTC). The tetrazolium salts INT and CTC are often used as markers of bacterial respiratory activity, as well as viability [20]. With these relatively simple methods, a good correlation between the number of INT/CTC-positive cells and the CFU count can be obtained.
Stay updated, free dental videos. Join our Telegram channel

VIDEdental - Online dental courses


