Introduction
Several candidate loci have been suggested as influencing mandibular prognathism (1p22.1, 1p22.2, 1p36, 3q26.2, 5p13-p12, 6q25, 11q22.2-q22.3, 12q23, 12q13.13, and 19p13.2). The goal of this study was to replicate these results in a well-characterized homogeneous sample set.
Methods
Thirty-three single nucleotide polymorphisms spanning all candidate regions were studied in 44 prognathic and 35 Class I subjects from the University of Pittsburgh School of Dental Medicine Dental Registry and DNA Repository. The 44 subjects with mandibular prognathism had an average age of 18.4 years; 31 were female and 13 male; and 24 were white, 15 African American, 2 Hispanic, and 3 Asian. The 36 Class I subjects had an average age of 17.6 years; 27 were female and 9 male; and 27 were white, 6 African American, 1 Hispanic, and 2 Asian. Skeletal mandibular prognathism diagnosis included cephalometric values indicative of Class III such as an ANB smaller than 2°, a negative Wits appraisal, and a positive A-B plane. Additional mandibular prognathism criteria included negative overjet and visually prognathic (concave) profile as determined by the subject’s clinical evaluation. Orthognathic subjects without jaw deformations were used as the comparison group. The mandibular prognathic and orthognathic subjects were matched by race, sex, and age. Genetic markers were tested by polymerase chain reaction with TaqMan chemistry. Chi-square and Fisher exact tests were used to determine overrepresentation of marker allele with an alpha of 0.05.
Results
An association was unveiled between a marker in MYO1H (rs10850110) and the mandibular prognathism phenotype ( P = 0.03). MYO1H is a Class I myosin that is in a different protein group than the myosin isoforms of muscle sarcomeres, which are the basis of skeletal muscle fiber typing. Class I myosins are necessary for cell motility, phagocytosis, and vesicle transport.
Conclusions
More strict clinical definitions might increase homogeneity and aid the studies of genetic susceptibility to malocclusions. We provide evidence that MYO1H can contribute to mandibular prognathism.
In orthodontics, one of the most challenging aspects in treating patients is predicting mandibular growth, especially in patients who show more pronounced characteristics of mandibular development. Through studies predominantly conducted with family members and twin siblings, it is well documented that there is a strong link between mandibular prognathism and genetics. More specifically, there is evidence of autosomal-dominant inheritance, with incomplete penetrance associated with this phenotype. The expression of the phenotype is a product of genetics and environmental factors. The multifactorial nature of mandibular prognathism makes it difficult to study and understand.
Mandibular prognathism has a prevalence as low as 1% in white people but as high as 15% in Asian populations.
Dohmoto et al found that in mice the size of the mandible was controlled by genes located at chromosomes 10 and 11 that correspond to human chromosomal regions 12q21 and 2p13, respectively.
In humans, genome-wide linkage analysis provided evidence for linkage to mandibular prognathism at chromosomes 1p36, 6q25, and 19p13.2. The presence of P56IT variant in the growth hormone receptor gene has been shown to affect mandibular morphology in Chinese and Japanese populations, especially in regard to mandibular height. In a study with a Hispanic cohort, Class III (due primarily to maxillary deficiency) was confirmed to be inherited in an autosomal-dominant pattern and linked to 5 loci: 1p22.1, 3q26.2, 11q22, 12q13.13, and 12q23. Li et al detected a specific locus, 14q24.3-31.2, associated with the mandibular prognathism phenotype in a Han Chinese population. In addition to the strong role of heredity, there has been evidence suggesting the contribution of certain environmental factors to mandibular prognathism such as enlarged tonsils, endocrine imbalances, posture, trauma, and disease.
It is well documented that the first studies of complex traits suggest a stronger genetic effect that was found by subsequent studies. Both bias and genuine population diversity might explain why early studies tended to overestimate the disease predisposition conferred by candidate gene polymorphisms. If there is a true effect of any of the previously described loci in mandibular prognathism, the expectation is that those results can be replicated. Therefore, we typed markers in 8 loci and measured the associations between genetic variations and mandibular prognathism using a population from Pittsburgh, Pennsylvania.
Material and methods
The subjects in this study were active orthodontic patients from the Department of Orthodontics of the School of Dental Medicine at the University of Pittsburgh. They were identified through the Dental Registry and DNA Repository project. In this project, since September 2006, people who seek treatment at the University of Pittsburgh School of Dental Medicine have been invited to be part of the registry. They provide written informed consent authorizing the extraction of information from their dental records. Also, they provide a saliva sample from which DNA can be extracted. Unstimulated saliva samples were obtained from all participants (they were asked to spit) and stored in Oragene DNA Self-Collection kits (DNA Genotek, Ottawa, Ontario, Canada) at room temperature until processing. No centrifugation was performed on the saliva samples. DNA was extracted according to the manufacturer’s instructions. This project was approved by the University of Pittsburgh Institutional Review Board.
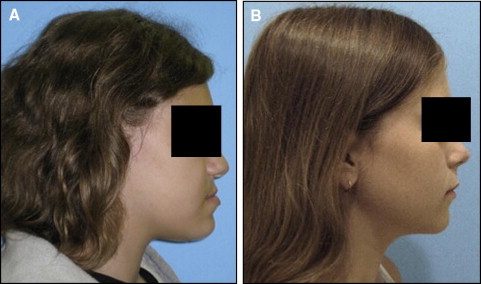
In September 2009, data from 1630 subjects were extracted from the registry for this pilot project. The 95 patients treated at the orthodontics department were considered eligible for this study. The assessment of the eligible subjects consisted of a careful review of each subject’s clinical and radiographic records. Clinical records consisted of digital orthodontic models (orthoCAD, Cadent, Carlstadt, NJ), digital tracings of lateral cephalograms (Dolphin, Chatsworth, Calif), and digital photographs. The first steps in the assessment process were to obtain all subjects’ soft-tissue profile photos (concave or straight profile) and certain cephalometric values to classify them as either orthognathic or prognathic. Specifically, we looked at Steiners’ ANB, Wits appraisal. and Downs’ A-B plane. As described in the Steiner analysis, ANB angles of less than 2° indicate that the mandible is located ahead of the maxilla. For this study, subjects with ANB values less than 2° were reviewed further to clarify whether the discrepancy was attributed to a smaller than average maxilla according to the SNA values. In that case, the subjects did not represent true prognathism but, rather, a regular-size mandible appearing to be protrusive because of a small maxilla. Therefore, they were excluded from the true prognathic group. The Wits appraisal was another measurement we reviewed because this value indicates the anteroposterior jaw relationship in the facial complex regardless of intracranial references. A negative Wits indicates a Class III skeletal relationship, and the more negative the value, the more severe the Class III. A Downs’ A-B plane angle larger than −4.6° indicates a skeletal Class III diagnosis, although this measurement might appear more severe in subjects with a pronounced bony pogonion. Additional Class III criteria included dental classifications, such as Class III molar and canine relationships, and negative overject based on the digital models and clinical examinations. We excluded subjects with facial clefting, abnormal anterior cranial base growth defects such as or similar to achondrodysplasia, or midfacial growth deficiencies caused by other pathologies such as tumors, cysts, or trauma that might scar midfacial periosteal surfaces and limit normal growth potentials. After analyzing the clinical information, we selected 44 mandibular prognathism and 36 orthognathic subjects for this study. The 44 mandibular prognathism subjects had an average age of 18.4 years; 31 were female and 13 male; and 24 were white, 15 African American, 2 Hispanic, and 3 Asian. The 36 Class I subjects had an average age of 17.6 years; 27 were female and 9 male; and 27 were white, 6 African American, 1 Hispanic, and 2 Asian. Table I describes all measurements used in the study. The Figure shows examples of the subjects in each comparison group.
| Profile | Sex | Age (y) | Ethnicity | Steiner’s ANB (°) | Wits appraisal (mm) | Downs’ A-B plane (°) |
|---|---|---|---|---|---|---|
| Prognathic | Female | 18 | White | −1.9 | −6.5 | 1.6 |
| Female | 20 | White | −4.9 | −6.8 | 6.6 | |
| Male | 17 | White | −8.1 | −12 | 12.7 | |
| Female | 23 | African American | −1.2 | −8.4 | 1.3 | |
| Female | 20 | White | 0.7 | −7.6 | −0.1 | |
| Female | 23 | White | −10.4 | −33.4 | 14.1 | |
| Female | 14 | Hispanic | −2.7 | −6.2 | 2.7 | |
| Female | 22 | White | −6.6 | −15.7 | 4.5 | |
| Female | 19 | African American | −3.4 | −6.4 | −2.8 | |
| Male | 19 | White | −1.1 | −7.3 | 1 | |
| Female | 30 | African American | −1.2 | −12.2 | 3.7 | |
| Female | 15 | African American | 2.4 | −3.4 | −2.4 | |
| Male | 15 | White | 0.3 | −1.3 | −1 | |
| Female | 16 | African American | −0.9 | −5 | 2.4 | |
| Female | 12 | Asian | −2.3 | −5 | 2.9 | |
| Female | 12 | Hispanic | 0.6 | −8.1 | 0.7 | |
| Female | 21 | African American | −1.9 | −4.5 | −1.7 | |
| Female | 13 | African American | −4 | −12 | 4.9 | |
| Female | 21 | African American | −4.8 | −4.3 | 4 | |
| Female | 15 | White | −7.5 | −13.3 | 9.1 | |
| Female | 28 | African American | −1 | −5.9 | 2.3 | |
| Female | 14 | White | −0.3 | −4.7 | 2 | |
| Male | 11 | White | −4.4 | −7.8 | 7.3 | |
| Female | 18 | White | −4.7 | −10.6 | 4.1 | |
| Male | 18 | White | −1.9 | −7 | 0.6 | |
| Female | 25 | White | −1.6 | −5.5 | 2.1 | |
| Male | 26 | White | −8.8 | −12.1 | 4.3 | |
| Female | 18 | African American | −6 | −10 | 7 | |
| Female | 11 | African American | −2.3 | −8.8 | 2.7 | |
| Female | 18 | White | −2.9 | −4.4 | 5.5 | |
| Male | 24 | Asian | −3.4 | −4.1 | 4.7 | |
| Female | 13 | White | −0.9 | −5.4 | −0.3 | |
| Female | 15 | White | −0.7 | −4.6 | 0.4 | |
| Female | 15 | African American | −1.2 | −7 | 2.6 | |
| Male | 22 | Asian | 0.3 | −6.6 | −1.7 | |
| Female | 15 | White | −3 | −10.6 | 3 | |
| Male | 28 | White | −1.9 | −8.3 | 2 | |
| Male | 12 | African American | −3.8 | −8.2 | 4.6 | |
| Male | 19 | White | −2.6 | −7.7 | 1.2 | |
| Male | 17 | African American | −2.2 | −3.6 | 1.8 | |
| Female | 13 | African American | −4.1 | −12.5 | 5.8 | |
| Male | 25 | White | −0.3 | −4.1 | −0.9 | |
| Female | 15 | White | −4.7 | −10 | 5.6 | |
| Female | 24 | White | −1.1 | −3.5 | 2.1 | |
| Orthognathic | Female | 14 | White | 1.8 | −0.2 | −3.4 |
| Female | 14 | White | 1.7 | 1.3 | −4.5 | |
| Female | 13 | African American | 2.4 | −0.9 | −2.7 | |
| Male | 14 | African American | 2.6 | −1.7 | −2.7 | |
| Female | 17 | White | 2 | −0.6 | −4.1 | |
| Male | 36 | White | 1.4 | −0.7 | −3.7 | |
| Male | 52 | White | 0.8 | −1.3 | −5.4 | |
| Female | 14 | White | 2.8 | −0.5 | −5.3 | |
| Female | 13 | White | 2.5 | 1.4 | −5 | |
| Female | 18 | White | 2.6 | −1.5 | −3.2 | |
| Male | 19 | White | 2.9 | −0.3 | −6.8 | |
| Female | 14 | White | 3.8 | 1.4 | −8.7 | |
| Female | 15 | White | 1.8 | 1 | −5 | |
| Male | 11 | African American | 2.3 | −2.9 | −3.4 | |
| Female | 24 | White | 2.4 | −2.7 | −3.7 | |
| Female | 15 | White | 0.1 | −2.7 | −0.3 | |
| Female | 15 | White | 2.5 | −3.2 | −2.5 | |
| Female | 15 | African American | 2.8 | −1.3 | −3.9 | |
| Female | 12 | White | 0.8 | −0.1 | −1.9 | |
| Female | 17 | White | 0.7 | −0.9 | −1.7 | |
| Male | 16 | White | 1.9 | −0.6 | −3.6 | |
| Female | 13 | White | 1.6 | −0.2 | −4.8 | |
| Female | 14 | White | 3.3 | 0.1 | −7.4 | |
| Female | 12 | White | 1.7 | 0.5 | −3.6 | |
| Female | 24 | White | 3.3 | 0.5 | −7.7 | |
| Male | 16 | White | 2.1 | −1.1 | −5.4 | |
| Female | 17 | Asian | 2.2 | −0.4 | −4.6 | |
| Female | 15 | White | 1.2 | −0.6 | −3.1 | |
| Female | 15 | White | 1 | −1.2 | −3.5 | |
| Female | 11 | Hispanic | 3.7 | 0.1 | −6 | |
| Female | 13 | White | 1.7 | 1.8 | −4.5 | |
| Male | 28 | Asian | 3.7 | 0.9 | −5.1 | |
| Female | 11 | African American | 1.9 | −1 | −1 | |
| Female | 11 | White | 3 | −0.1 | −4.6 | |
| Female | 16 | African American | 3.8 | 0.1 | −4.8 | |
| Male | 22 | White | 2.7 | −4 | −2.6 |
We selected 33 single nucleotide polymorphisms covering the 8 candidate regions studied ( Table II ) from the International HapMap Project database ( http://www.hapmap.org ). We used the function “Download tag SNP data” and selected 26 polymorphisms as representative of the polymorphisms in the region. We selected polymorphisms that maximally represented the linkage disequilibrium structure of a region to avoid redundant information. Preference was given to polymorphisms with high heterozygosity levels and different minor allele frequencies to avoid intermarker linkage disequilibrium.
| Marker | Locus | Gene | Original study suggesting as a candidate region |
|---|---|---|---|
| rs2503243 | 1p22.2 | Intergenic | Frazier-Bowers et al, 2009 |
| rs972054 | 1p22.2 | Intergenic | Frazier-Bowers et al, 2009 |
| rs1413533 | 1p22.2 | Intergenic | Frazier-Bowers et al, 2009 |
| rs4649030 | 1p36.11 | Intergenic | Yamaguchi et al, 2005 |
| rs2101560 | 3q26.32 | Intergenic | Frazier-Bowers et al, 2009 |
| rs1490055 | 3q26.32 | Intergenic | Frazier-Bowers et al, 2009 |
| rs2087312 | 3q26.32 | Intergenic | Frazier-Bowers et al, 2009 |
| rs987526 | 3q26.32 | Intergenic | Frazier-Bowers et al, 2009 |
| rs1601948 | 3q26.32 | Intergenic | Frazier-Bowers et al, 2009 |
| rs1387168 | 3q26.32 | Intergenic | Frazier-Bowers et al, 2009 |
| rs2940913 | 5p12 | Growth hormone receptor ( GHR ) | Yamaguchi et al, 2001 ; Zhou et al, 2005 ; Kang et al, 2009 ; Tomayasu et al, 2009 |
| rs2973015 | 5p12 | Growth hormone receptor ( GHR ) | Yamaguchi et al, 2001 ; Zhou et al, 2005 ; Kang et al, 2009 ; Tomayasu et al, 2009 |
| rs1509460 | 5p12 | Growth hormone receptor ( GHR ) | Yamaguchi et al, 2001 ; Zhou et al, 2005 ; Kang et al, 2009 ; Tomayasu et al, 2009 |
| rs2910875 | 5p12 | Growth hormone receptor ( GHR ) | Yamaguchi et al, 2001 ; Zhou et al, 2005 ; Kang et al, 2009 ; Tomayasu et al, 2009 |
| rs7718944 | 5p12 | Growth hormone receptor ( GHR ) | Yamaguchi et al, 2001 ; Zhou et al, 2005 ; Kang et al, 2009 ; Tomayasu et al, 2009 |
| rs7750085 | 6q26 | Parkinson juvenile disease protein 2 ( PARK2 ) | Yamaguchi et al, 2005 |
| rs3016534 | 6q26 | Parkinson juvenile disease protein 2 ( PARK2 ) | Yamaguchi et al, 2005 |
| rs12207168 | 6q26 | Parkinson juvenile disease protein 2 ( PARK2 ) | Yamaguchi et al, 2005 |
| rs9458378 | 6q26 | Parkinson juvenile disease protein 2 ( PARK2 ) | Yamaguchi et al, 2005 |
| rs1884153 | 6q26 | Parkinson Juvenile disease protein 2 ( PARK2 ) | Yamaguchi et al, 2005 |
| rs571407 | 11q22.3 | Caspase 4 isoform gamma precursor ( CASP4 ) | Frazier-Bowers et al, 2009 |
| rs1902768 | 12q13.13 | Keratin 7 ( KRT7 ) | Frazier-Bowers et al, 2009 |
| rs7300317 | 12q13.13 | Keratin 7 ( KRT7 ) | Frazier-Bowers et al, 2009 |
| rs11113231 | 12q23.3 | Intergenic | Frazier-Bowers et al, 2009 |
| rs4964541 | 12q23.3 | Intergenic | Frazier-Bowers et al, 2009 |
| rs10850110 | 12q24.11 ∗ | Myosin 1H ( MYO1H ) | Frazier-Bowers et al, 2009 |
| rs10850364 | 12q24.21 ∗ | T-box 3 ( TBX3 ) | Frazier-Bowers et al, 2009 |
| rs7351083 | 19p13.2 | Fibrilin 3 precursor ( FBN3 ) | Yamaguchi et al, 2005 |
| rs4804264 | 19p13.2 | Fibrilin 3 precursor ( FBN3 ) | Yamaguchi et al, 2005 |
| rs8103218 | 19p13.2 | Fibrilin 3 precursor ( FBN3 ) | Yamaguchi et al, 2005 |
| rs12327845 | 19p13.2 | Fibrilin 3 precursor ( FBN3 ) | Yamaguchi et al, 2005 |
| rs10411185 | 19p13.3 | Intergenic | Yamaguchi et al, 2005 |
Stay updated, free dental videos. Join our Telegram channel

VIDEdental - Online dental courses


