Gene organization and expression in eukaryotes
Publisher Summary
This chapter discusses gene organization and expression in eukaryotes. There are a number of fundamental differences between the way genes are arranged, expressed, and controlled in eukaryotic cells when compared with bacteria. Eukaryotes, because of their greater complexity, must possess much more genetic information. Primary RNA transcripts in eukaryotic cells are extensively modified, cleaved, and spliced in the nucleus before being transported to the cytoplasm in the form of mRNA. The control of gene expression is much more complex and diverse in eukaryotes, with many levels of regulation. In addition to transcriptional control, posttranscriptional mechanisms also play a major role. The primary translation products may, in turn, be subjected to posttranslational modification. DNA replication in eukaryotic cells is fundamentally the same as that in bacteria. Replication is semi-conservative, occurs in a 5’ → 3’ direction, and requires a template, a primer, and deoxyribonucleoside triphosphates for synthesis to occur. Replication also precedes bidirectionally, but in view of the large size of eukaryotic chromosomes, synthesis is initiated simultaneously at many origins; otherwise chromosome replication would take many days.
Genes and chromosomes
1. Eukaryotes, because of their greater complexity, must possess much more genetic information. For example, a human cell contains about 1000 times more DNA than an E. coli cell. As a result the DNA may have an overall length of several centimetres which must be packed into a few micrometres. Hence the DNA of eukaryotic cells is very highly condensed and this is aided by the presence of a class of basic proteins called histories. Bacterial DNA is not associated with histones.
2. Eukaryotic chromosomes are located within a nucleus bounded by a nuclear membrane and, since proteins are synthesized in the cytoplasm, the sites of transcription and translation are physically separated. Consequently these two processes are not as closely coupled as they are in bacteria.
3. Primary RNA transcripts in eukaryotic cells are extensively modified, cleaved and spliced in the nucleus before being transported to the cytoplasm in the form of mRNA.
4. The control of gene expression is much more complex and diverse in eukaryotes, with many levels of regulation. In addition to transcriptional control, post-transcriptional mechanisms also play a major role. Furthermore, the primary translation products may in turn be subjected to post-translational modification.
Eukaryotic chromosome structure
Nucleosomes
Several lines of evidence have shown that each nucleosome is composed of a core particle containing 140 base pairs of DNA (Figure 21.1), irrespective of the species or cell type. This DNA is wound in the form of 1·75 turns of a left-handed superhelix, round a histone octamer composed of two molecules each of H2A, H2B, H3 and H4. Adjacent nucleosomes are then joined by a short stretch of DNA, called linker DNA, thereby maintaining the integrity of the chromosome. Consequently a typical mammalian gene of 10000 base pairs will contain about 40 nucleosomes. Histone H1, which is present at only one molecule per nucleosome and is absent from the core particle, may be associated with the linker DNA and serve as a bridge between adjacent nucleosomes.
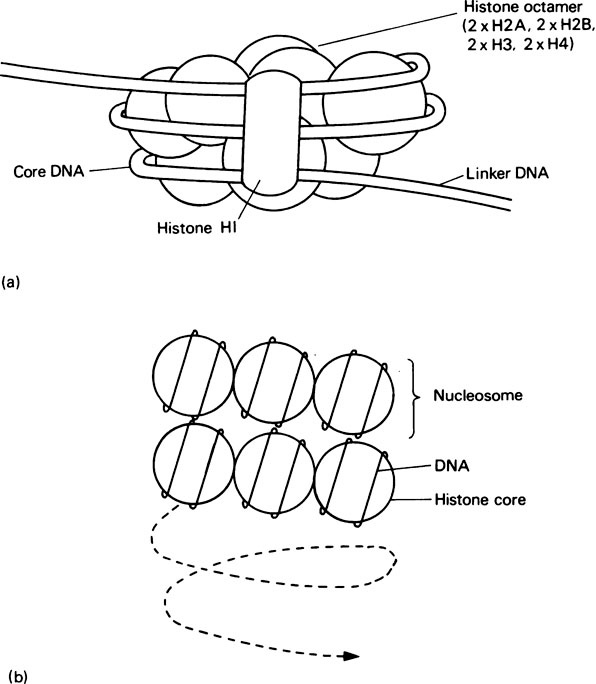
If stretched out a 200 base pair piece of DNA (140 base pairs in a nucleosome core plus about 60 base pairs of linker DNA) would have a length of 680 Å (68 nm). This is packed within a 100 Å (10 nm) diameter nucleosome by winding round the outside. Hence the packing ratio of a nucleosome is about 7. However, the overall packing ratio of DNA in interphase nuclei is about 100−1000 and this is further increased to 10000 during metaphase. Consequently the formation of nucleosomes must be only the first step in DNA packing. At the moment it is not certain what other higher levels of chromatin packing exist. However, one popular model is the solenoid model. This proposes that chromatin forms a solenoid with six nucleosomes to the turn of the helix (Figure 21.1) giving an overall structure with a diameter of about 360 Å (36 nm) and a packing ratio of 40. Further folding of the solenoid into loops would then result in an even more condensed structure with a packing ratio approaching that found in interphase chromosomes. A class of nuclear proteins called the non-histone proteins may play an important part in stabilizing such postulated higher levels of chromatin structure.
Stay updated, free dental videos. Join our Telegram channel

VIDEdental - Online dental courses


